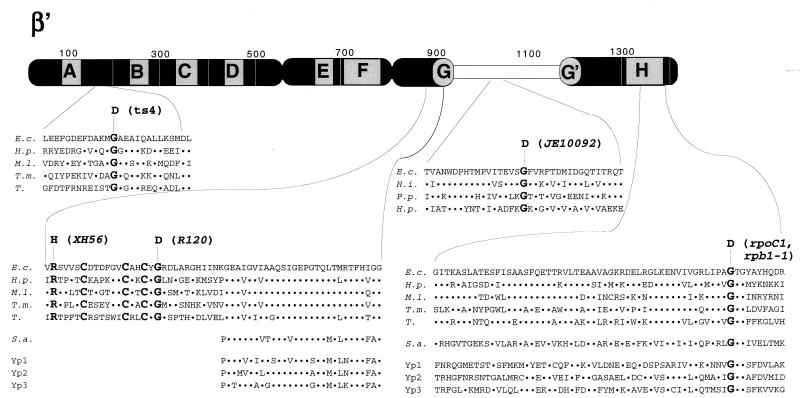
FIG. 2.
Genetic context of rpoC assembly mutants. The thick bar represents the 1,407-amino-acid β′ subunit of E. coli RNAP. Stippled boxes labeled A to H represent segments of β′ highly conserved in evolution (1, 24). The E. coli hypervariable region (24), absent in homologues from most bacteria, archaea, and eukaryotes, is shown by an open box. The amino acid sequences of E. coli β′ (E.c.) harboring assembly mutations are expanded underneath. Homologous amino acid sequences from Haemophilus influenzae (H.i.), Pseudomonas putida (P.p.), Helicobacter pylori (H.p.), Mycobacterium leprae (M.l.), Thermotoga maritima (T.m.), chloroplasts from tobacco (T.), Sulfolobus acidocaldarium (S.a.), and yeast RNAP 1, 2, and 3 (Yp1, Yp2, and Yp3, respectively) are also shown. The dots symbolize identity to the E. coli sequence. Amino acids changed by assembly mutations, as well as three conserved cysteines close to segment G, are highlighted by bold typeface. Note that rpoC1 in E. coli and rpb1-1 in yeast cause the same substitution.