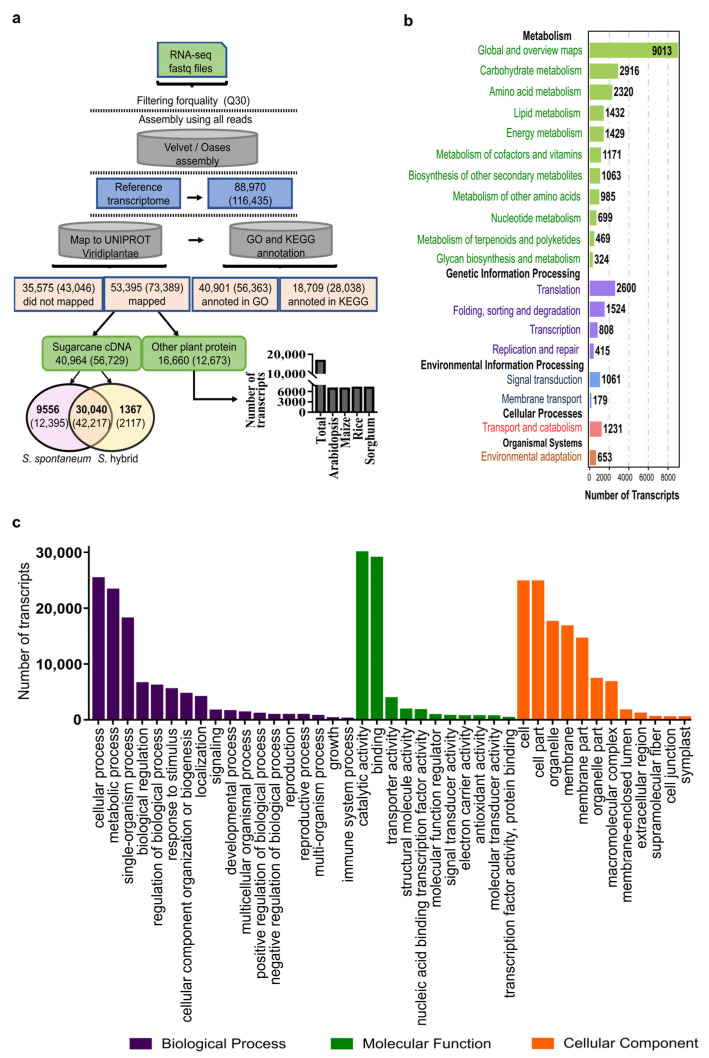
Figure 1.
Pipeline of the construction of the sugarcane reference transcriptome. (a) A total of 16 libraries were analyzed and sequencing results were used to construct a sugarcane reference transcriptome. Over 130 million reads were used for Velvet-Oases pipeline assembly that resulted in 88,970 loci and 116,435 transcripts. A total of 53,395 loci mapped with plant sequence databases and, within these, 40,964 matched with sugarcane genome sequences available in public databases. A group of 12,673 loci mapped with other plant species databases. A group of 35,575 loci did not map within Viridiplantae databases. The figure also shows the number of sugarcane transcripts with no matches in sugarcane ESTs database that presented homologues in Arabidopsis, maize, rice and sorghum with an e-value cut-off < 10−5, over 70% of identity and best hits. The transcripts were categorized in (b) KEGG pathway annotation and (c) GO database, using OmicShare tools. Only the most represented classes in sugarcane reference transcriptome were displayed.