Extended Data Fig. 5. Structural analysis of the interaction between UvrD-CTD and UvrB.
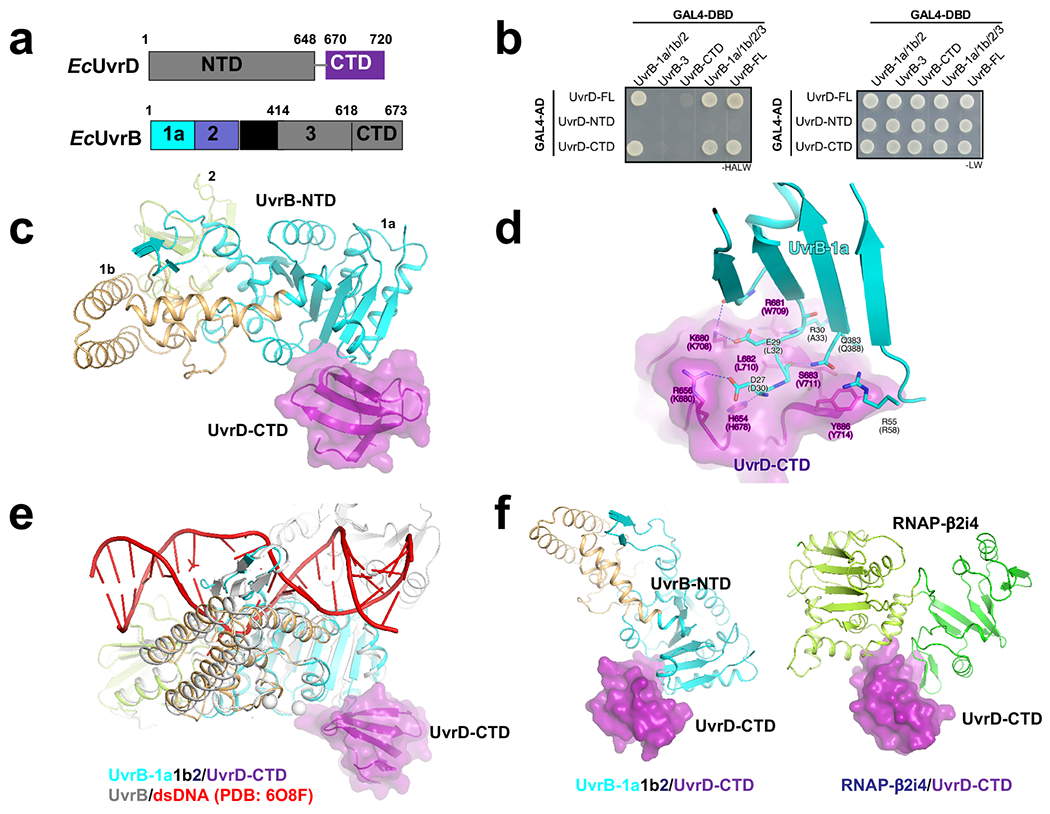
a, UvrD and UvrB domains (numbered as in E. coli). The domains observed in crystal structure are highlighted in colors. b, UvrD-CTD interacts with UvrB-1a/1b/2domain (or UvrD-NTD), consistent with a previous report34. E. coli UvrD (full-length or truncated) and UvrB (full-length or truncated) were fused to GAL4-AD and GAL4-DBD, respectively. The potential interactions were selected on SD (-HALW) plates, and the growth on SD (-LW) plates was used as input control. c, A 2.6-Å crystal structure of T. thermophilus UvrD-CTD/UvrB-NTD complex (PDB 7EGT). UvrB-1a docks on a shallow groove of UvrD-CTD. UvrD-CTD, UvrB-1a, UvrB-1b, and UvrB-2 are colored in purple, cyan, orange, and light green, respectively. d, The detailed interaction between UvrD-CTD and UvrB-1a. Residues H654, R656, K680, R681, S683 of UvrD-CTD makes a H-bond network with residues D27, E29, R30, Q383 of UvrB-1a (residues are labeled and numbered as in T. thermophilus; the corresponding residues in E. coli are indicated in parentheses). Y686 of UvrD-CTD makes stacking interaction with R55 and Q383 of UvrB-1a. e, Structural superimposition of UvrB-NTD/UvrD-CTD complex (colored as above) and UvrB/dsDNA complex (gray and red; PDB 6O8F)61 shows that UvrD-CTD binds the opposite surface of UvrB dsDNA-loading cleft, implicating UvrD doesn’t affect dsDNA loading of UvrB. f, Structural comparison between UvrD-CTD/UvrB-NTD (left) and UvrD-CTD/RNAP-β2i4 (right, PDB 7EGS) shows that UvrB and RNAP-β2i4 binds at the same cleft of UvrD-CTD, and thereby suggests that the interactions of UvrB and RNAP to UvrD are mutually exclusive.
