Extended Data Fig. 11. Local transcription enables NER (extension to Fig. 5).
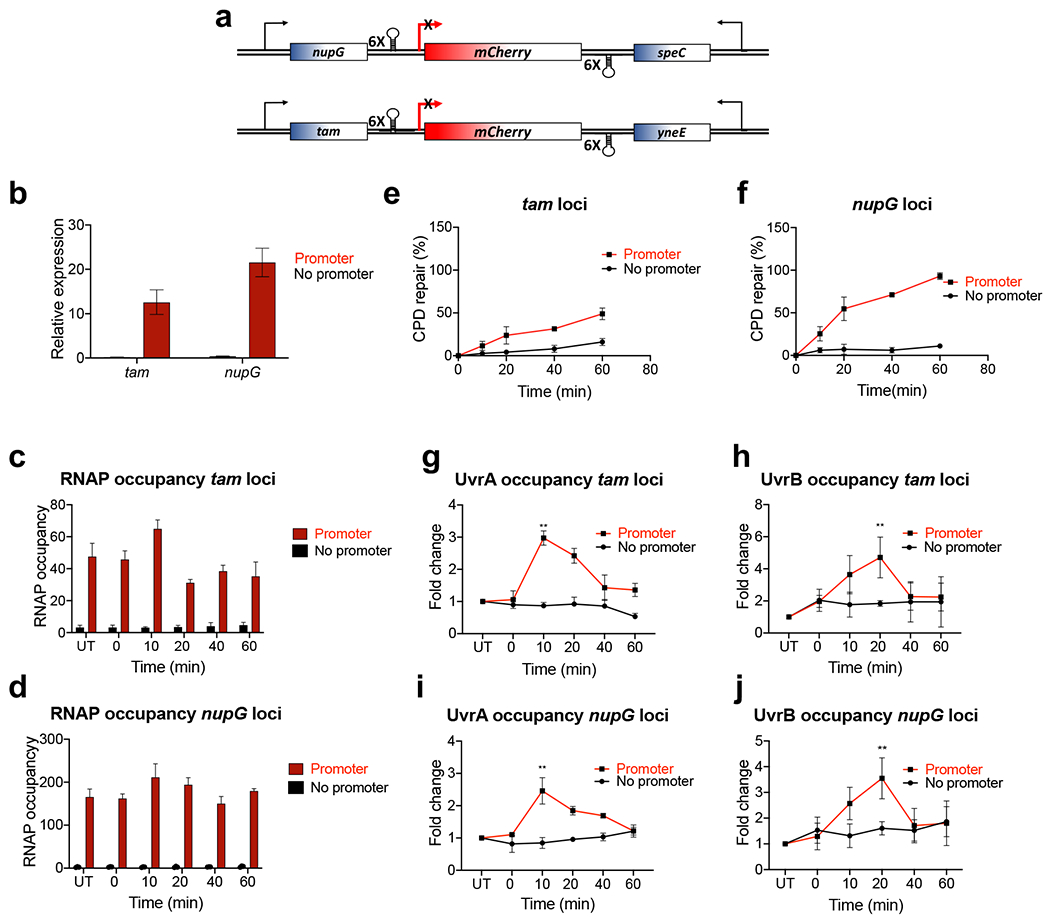
a-d, Depriving the genomic loci of transcription abolishes their NER. a, Schematics of the mCherry insulators. A transcription unit containing mCherry (with or without lacZ promoter) flanked by the terminator cassettes was inserted at the tam and nupG loci. b-d The expression of mCherry from the insulators (b) and RNAP occupancy (c, d) upon IPTG induction, as determined by RT-qPCR and ChIP-qPCR, respectively. Values are the means ± SD (n = 3). e,f CPD repair within the insulators. Most of NER strictly depends on promoter-initiated transcription. The levels of transcription and NER are stronger within the nupG insulator comparing to the tam insulator. Cells were induced with IPTG followed by UV irradiation (40 J/m2) and recovery in the dark for the indicated time intervals. CPD density was determined by SLR-qPCR as in Fig. 5a and used to calculate the percentage of repaired CPDs. Values are the means ± SD (n = 3). g-j, UvrAB recruitment to the UV-damaged DNA strictly depends on local transcription in both tam and nupG loci. Occupancy of RNAP (c,d), UvrA (g,i) and UvrB (h,j) following UV irradiation was determined by ChIP-qPCR. Cells were induced with IPTG followed by UV irradiation (40 J/m2) and recovery for the indicated time intervals. Values are the means ± SD (n = 3). Results are shown as a fold change in the occupancy of UvrAB within the insulator following UV irradiation. UT – untreated. Values are means ± SD (n = 3). **P < 0.01, ****P < 0.0001 (Student’s t test; equal variance). P values compare the percentage of DNA repair between “promoter” and “no promoter” strains for a given time point.
