Fig. 2. Structural organization of the pre-TCR and TCR complexes in cellulo.
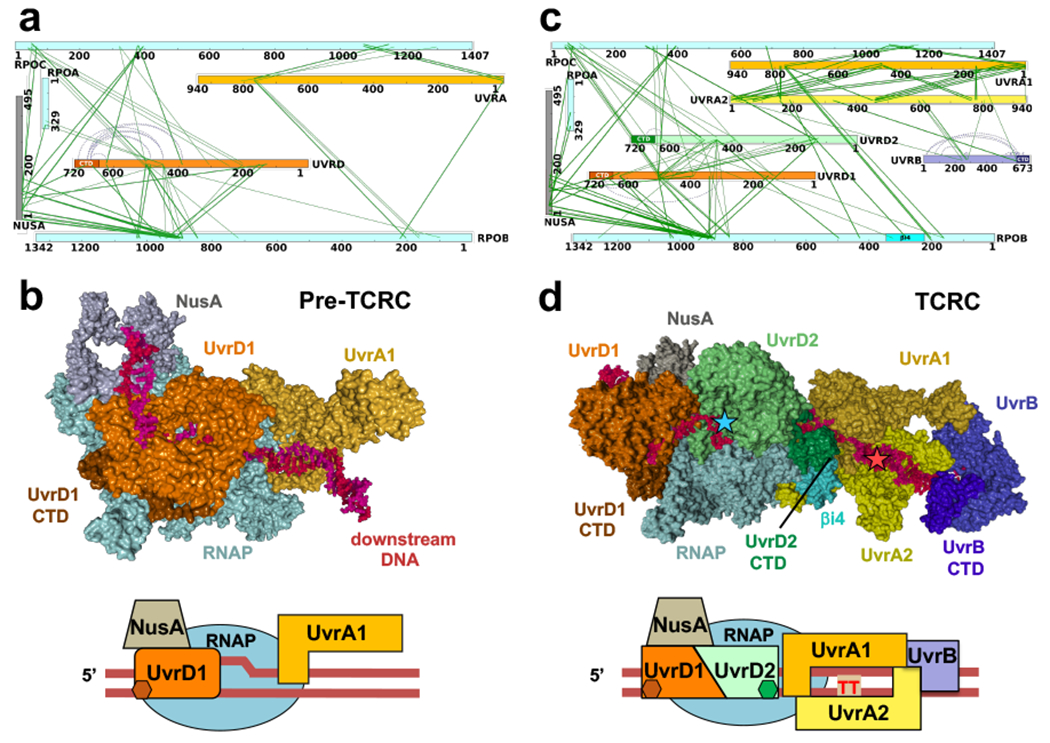
a,b, Architecture of the pre-TCRC based on in vivo and in vitro XLMS. a, Network view of highly confident non-redundant inter-protein crosslinks between RNAP subunits, NusA, UvrA, and UvrD. In vivo crosslinks were generated prior to genotoxic stress. Crosslinks were aggregated from DSS and EDC datasets (Supplementary Table 1). b, XLMS-driven pre-TCRC model. The model was built by crosslink-guided docking using PatchDock and the workflow described in Extended Data Fig. 18 and Methods. The cartoon shows the positioning of UvrAD monomers relative to the nucleic acid scaffold and RNAP. CTD of UvrD1 is shown as a brown hexagon. RNA is not shown. c,d, Architecture of the TCRC based on in vivo XLMS. c, Network view of highly confident non-redundant inter-protein crosslinks between RNAP subunits, NusA, UvrA, UvrB, and UvrD. Crosslinks were aggregated from in vitro DSS, in vivo DSS, and EDC datasets generated 20 min after 4NQO treatment (Supplementary Table 1). d, XLMS-driven TCRC model (see also Movie 1). The model was built by crosslink-guided docking using PatchDock and the workflow described in Extended Data Figs. 18, 19, and Methods. The cartoon shows the positioning of UvrABD monomers relative to the nucleic acid scaffold and a hypothetical CPD lesion (TT) in the backtracked TCRC. CTD of UvrD2 is shown as green hexagon. RNA is not shown.
