Extended Data Fig. 18. An outline of the automated workflow for crosslink-guided docking.
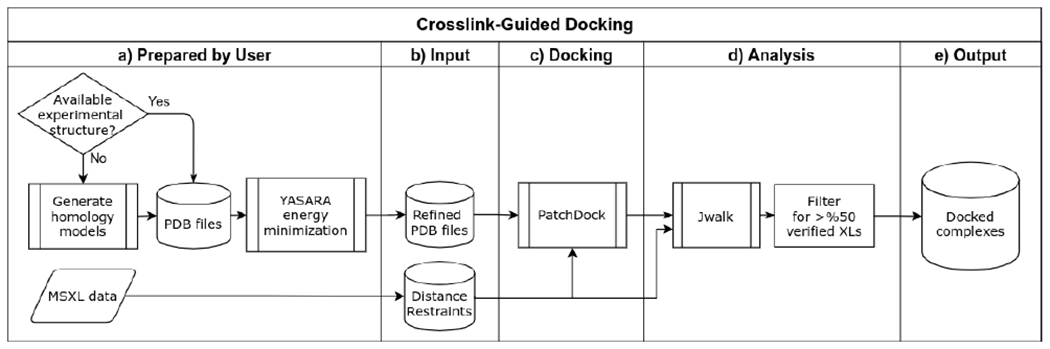
a,b, Coordinate files for all the E. coli interactors used to build the pre-TCRC and TCRC are prepared using available PDB structures, which were refined using YASARA Structure67 (see Methods). Proteins without available PDB structures were homology-modeled using I-TASSER68,69. XLMS data was converted to the distance restrains compatible with PatchDock. c, PDB files of receptor and ligand molecules were submitted to PatchDock with their corresponding distance restrains for rigid-body docking. d, The docking results were validated by examining the crosslink satisfaction using Jwalk70 (see Methods).
