Extended Data Fig. 20. Application of the docking pipeline to model the pre-TCRC.
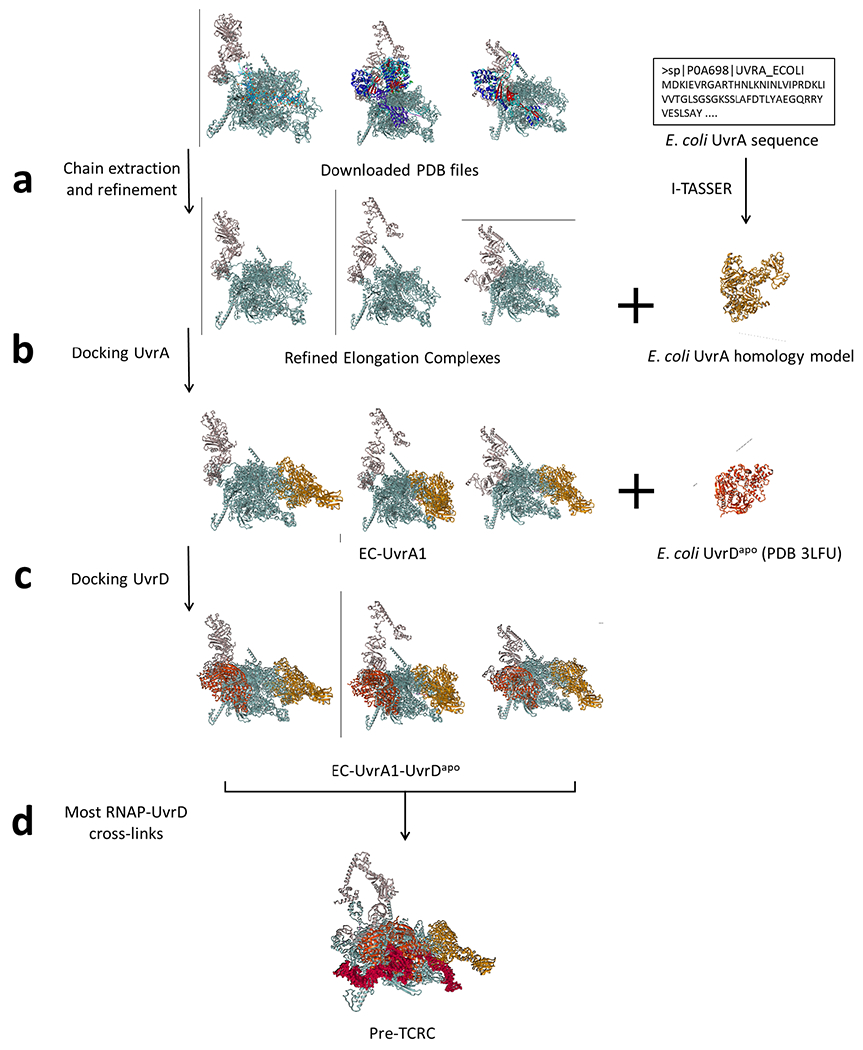
a, PDB files of E. coli elongation complexes were downloaded and prepared by extracting chains corresponding to RNAP subunits and NusA, then refined using the energy-minimization protocol included in YASARA Structure. E. coli UvrA was modeled using the homology template server I-TASSER. b, UvrA (ligand) was docked to ECs (receptors) from the previous step using PatchDock, with the RNAP-UvrA crosslinks provided as distance restraints. c, The PDB coordinates file of E. coli UvrD in the apo form was trimmed to the first 640 residues and refined using YASARA Structure, then docked to the top EC-UvrA complexes generated in the previous step, as ranked by RNAP-UvrA crosslink satisfaction. d, EC-UvrA-UvrD complexes generated in the previous step were ranked by RNAP-UvrD crosslink satisfaction and used as receptors to dock UvrD-CTD. Results were clustered using ProFit (V3.1), and finally analyzed for alignment of UvrAD DNA-binding regions with the DNA path in the EC.
