Extended Data Fig. 3. Reconstitution of the TCRC without nucleic acids.
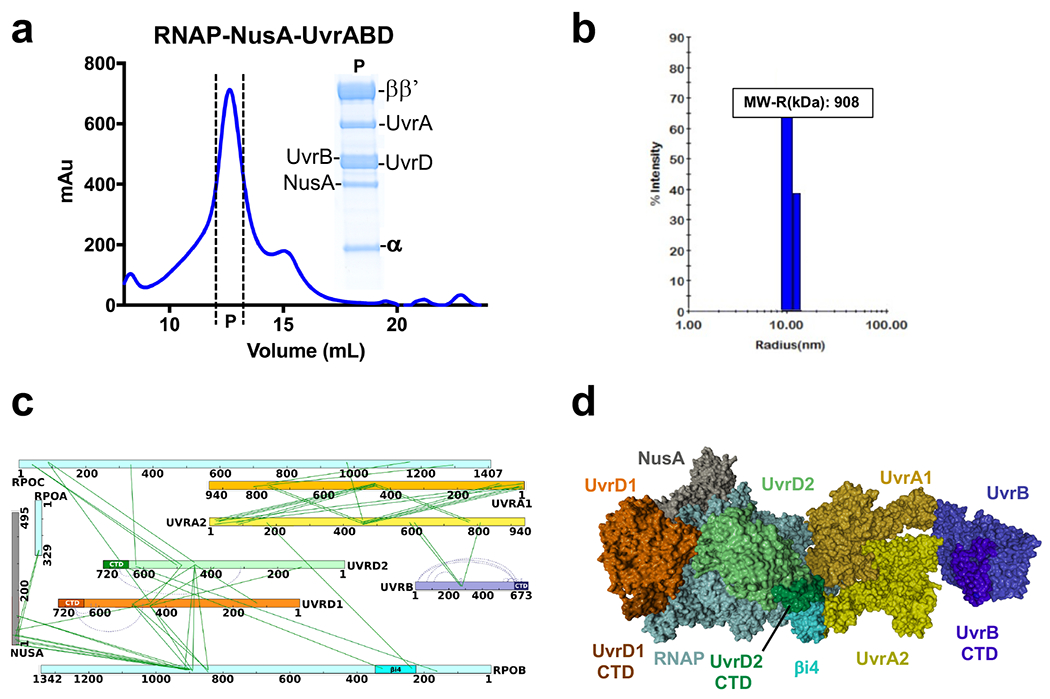
a, Isolation of the RNAP:NusA:UvrABD complex by SEC. SDS-Coomassie gel represents the protein fraction eluted from the main peak (P). b, DLS analysis of the RNAP:NusA:UvrABD complex. “P” fraction from (a) was subjected to DLS. Raleigh sphere (R) estimate of the complex molecular weight (MW = 908 kD), which deviates by only 1.7% from the theoretical MW of a uniform monodispersed complex containing 1 RNAP, 1 NusA, 2 UvrD, 2 UvrA, and 1 UvrB molecules, c, Network view of the highly confident non-redundant inter-protein crosslinks. Crosslinks were aggregated from DSS datasets (Supplementary Table 1). d, XLMS-based model of the reconstituted RNAP:NusA:UvrABD complex. The model was built based on the in vitro crosslinks using PatchDock and the workflow described in Extended Data Figs. 18, 19, and Methods.
