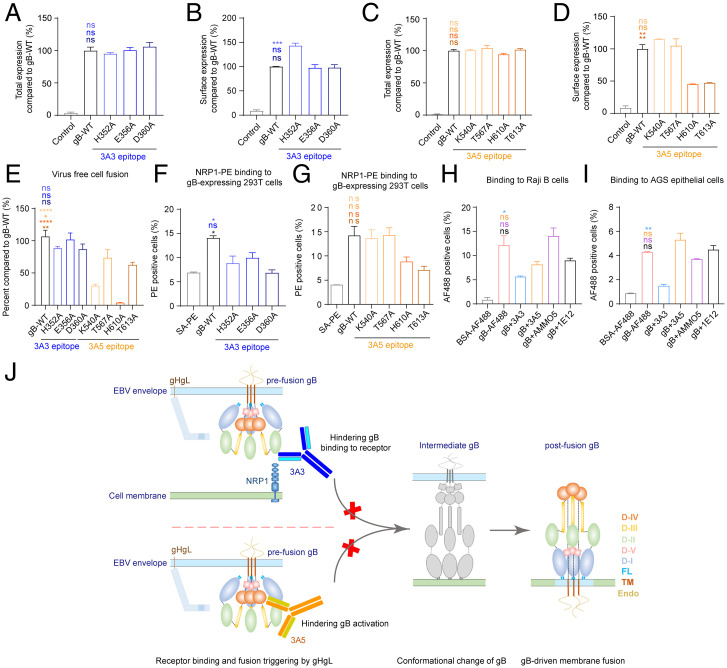
Fig. 7.
3A3 and 3A5 binding sites at gB D-II and D-IV are critical for cell binding and fusion. (A-D) The total expression and cell surface expression levels of gB-WT and gB 3A3 mutants (A and B) or 3A5 mutants (C and D) produced by transfected 293T cells were measured with mAb 3A5 (A and B) and mAb 3A3 (C and D) by flow cytometry, respectively. Antibody staining was performed with cell membrane permeabilization when the total expression was evaluated in A and C. Antibody staining was performed without cell membrane permeabilization when cell surface expression was evaluated in B and D. All values were normalized as a percentage to gB-WT. (E) Cell-cell fusion efficiency of gB-WT and gB mutants. All values were normalized as a percentage to gB-WT. (F and G) The binding of NRP1 to 293T cells expressing gB-WT, 3A3 epitope mutants (F), or 3A5 epitope mutants (G) of gB was evaluated by flow cytometry without cell membrane permeabilization. Cells stained with SA-PE alone were used as a negative control. (H and I) The binding of gB to Raji B cells (H) and AGS gastric adenocarcinoma epithelial cells (I) in the presence of anti-gB antibodies was evaluated by flow cytometry without cell membrane permeabilization. Bovine serum albumin (BSA)-AF488 and gB-AF488 were used as negative and positive controls, respectively. (J) Proposed schematic diagram of neutralizing mechanisms of 3A3 and 3A5. 3A3 blocks gB binding to its coreceptor NRP1 by directly restricting access to the interface. The binding of 3A5 to D-IV could restrict the movement of the gB trimer during conformational changes from pre- to postfusion by steric hindrance. It is also possible that the binding of 3A3 and 3A5 could bring steric hindrance that inhibits the triggering of gB activation by gHgL. Data are represented as the mean of two independent replicates ± SEMs. Statistical analyses were performed using one-way ANOVA. The color of the asterisks denotes the group with which there is a significant difference determined by a Sidak multiple comparison test. ns, no significant difference. *P ≤ 0.0332; **P ≤ 0.0021; ***P ≤ 0.0002; ****P ≤ 0.0001. SA-PE, streptavidin-phycoerythrin. PE, phycoerythrin. FL, fusion loop. TM, transmembrane domain. Endo, endodomain.