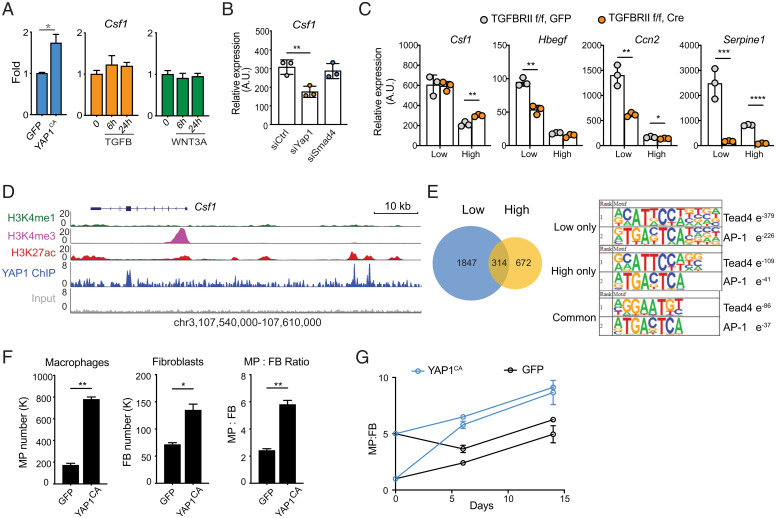
Fig. 4.
YAP1-dependent regulation of Csf1 in fibroblasts controls macrophage numbers. (A) Csf1 expression in MEFs carrying YapCA (Left), or treated with recombinant TGF-β (Center) or WNT3A (Right). MEFs were isolated from YapKIfl/fl mice and then transduced with lentivirus carrying GFP or Cre-GFP (constitutively-active, YAPCA). (B) Csf1 expression in MEFs 3 d after transfection with siRNAs targeting Yap1, Smad4, or scrambled control siRNA (siCTRL). (C) Expression of selected genes in wild-type and Tgfbr2 knockout (KO) MEFs cultured at low or high density. Wild-type and Tgfbr2 KO MEFs are generated by viral transduction of Tgfbr2fl/fl MEFs with GFP or Cre-GFP vectors, respectively. (D) Genomic tracks displaying ChIP-seq occupancy of histone modifications (H3K27ac, H3K4me1, and H3K4me3) and endogenous YAP1 binding at the Csf1 gene locus in MEFs. (E) Venn diagram showing the number of YAP1 binding peaks at low and high cell densities determined by ChIP-seq. The top ranked enrichment of transcription factor motifs is shown for peaks at high, low, or both cell densities. (F and G) Wild-type (GFP) and YapCA MEFs were plated together with WT BMDMs. Their numbers and ratios were determined by flow cytometry 6 d after coculture (F). BMDMs and MEFs were plated at different starting ratios and quantified at days 6 and 14 (G) *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.