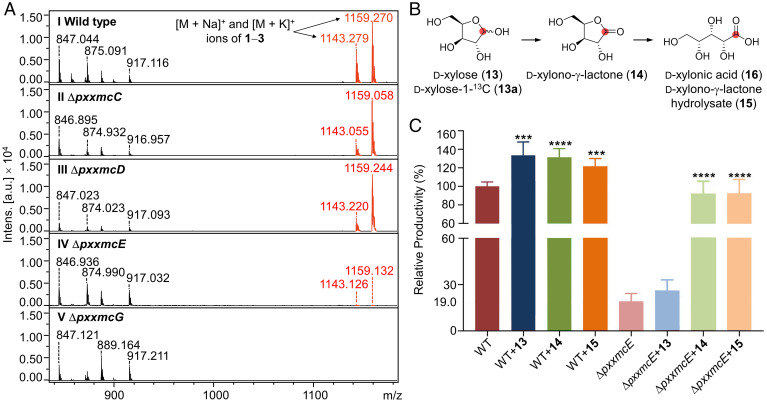
Fig. 4.
Functional analysis of pxxmc biosynthetic genes. (A) MALDI-TOF MS traces of extracts from (I) Paramyrothecium sp. XJ0827 wild-type or (II to V) mutant strains. All strains were cultivated for 10 d on PDA media. Ions with m/z of 847, 875, 889, and 917 derive from unrelated metabolites. They show little variation in their intensities (Intens.) among the various strains, illustrating that the ΔpxxmcE and ΔpxxmcG mutations specifically affect the productivities of xylomyrocins 1–3. (B) Proposed conversion of d-xylose (13) or d-xylose-1-13C (13a) to d-xylonic acid (16). The position of the label in 13a is indicated with a red dot. (C) Relative xylomyrocin productivities of the wild-type and ΔpxxmcE strains upon supplementation with d-xylose (13), d-xylono-γ-lactone (14), and d-xylono-γ-lactone hydrolysate (15). Hydrolysate 15 was prepared by treating 14 with 1 M aqueous NaOH solution. Both strains were cultivated for 6 d in liquid M-100 media before supplementation of 13, 14, or 15 at 3 mM (final concentration). Fermentation was continued for an additional 3 d to facilitate complete utilization of the supplemented substrate. Data were collected from three independent experiments of three replicates each (n = 9). Columns represent the mean ± SD. ***P < 0.001 and ****P < 0.0001 as determined with unpaired Student two-tailed t tests comparing the yields of the substrate-supplemented fermentations with that of the nonsupplemented fermentation. a.u., arbitrary units.