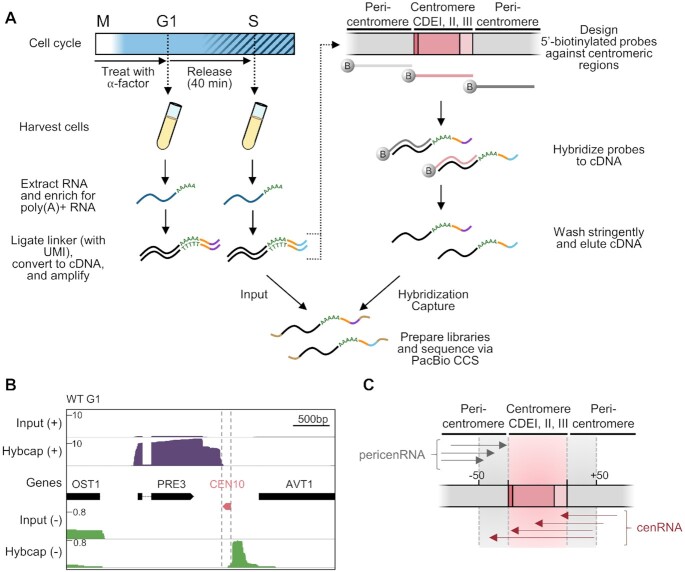
Figure 1.
Hybridization capture and long-read sequencing of lowly expressed cenRNAs. (A) Schematic illustrating the procedure to capture and sequence RNA molecules generated at or near budding yeast centromeres. Cells were synchronized in G1 phase with α-factor and then released into the cell cycle. Cells were harvested immediately after release (G1 phase) and 40 min after release (S phase). RNA (dark blue) was extracted and enriched for those that were polyadenylated. Poly(A)+ RNA was ligated to a linker (orange) containing a unique molecular identifier (UMI; purple or light blue). The RNA was next reverse transcribed into double-stranded cDNA (black) and amplified via PCR. The cDNA was hybridized to 5′-biotinylated DNA probes (dark grey, light grey and pink) designed against the three CDEs (pink) of all 16 centromeres as well as the flanking regions (gray) to enrich for centromeric and pericentromeric RNAs. The probes were washed stringently and the remaining cDNA was eluted from the probes. Both input and enriched (Hybcap) cDNAs were prepared into a library by adding 5′ and 3′ barcoded adapters (brown) and sequenced via Pacific Biosciences (PacBio) long-read circular-consensus sequencing (CCS). (B) Iso-Seq profiles of WT G1 cells before (Input) and after hybridization capture (Hybcap). Position of centromere boundaries is shown by the vertical grey dotted lines. Reads coming from the (+) strand are shown in purple while reads coming from the (–) strand are shown in green. (C) Schematic illustrating the nomenclature used throughout the paper. PericenRNAs correspond to transcripts converging towards the CEN but stopping within 50 bp of the CEN border (grey arrows) while cenRNAs correspond to transcripts that converge towards the CEN and enter it by at least 1bp (dark pink arrows). cenRNAs are pericenRNAs that readthrough the CEN.