Figure 1.
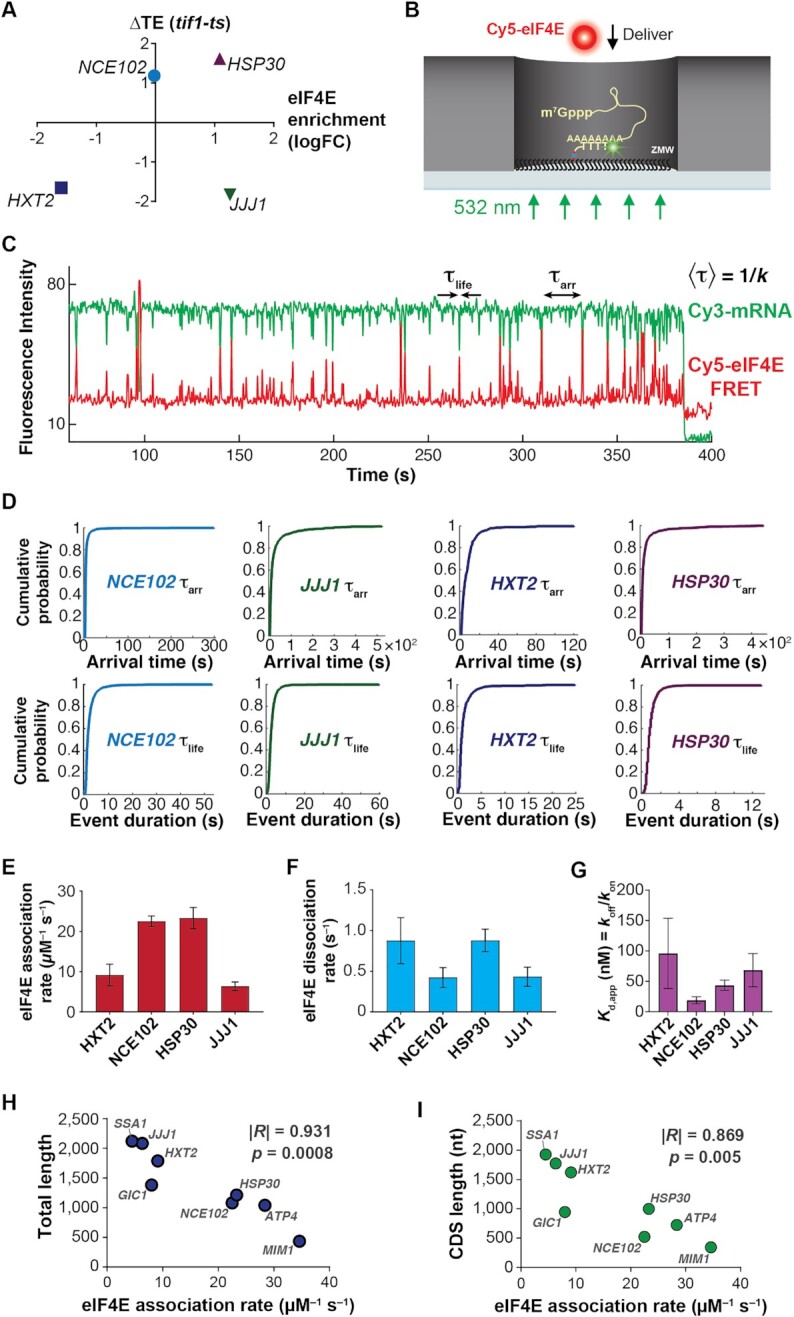
eIF4E interaction dynamics with full-length mRNAs depend on mRNA identity and length. (A) Selection of mRNAs with varying in-vivo enrichment in eIF4E•eIF4G and translation dependence on eIF4A, as measured by Costello et al. (2015), and Sen et al. (2015). (B) Schematic of single-molecule FRET experiment to detect binding of fluorescently-labeled eIF4E to surface-immobilized, fluorescently-labeled mRNA. (C) Sample smFRET trajectory showing eIF4E–mRNA interaction in the absence of other eIF4F components. (D) Representative cumulative distribution functions for eIF4E association with (top) and dissociation from (bottom) full-length mRNAs. (E) eIF4E–mRNA association rates quantified from exponential fitting of arrival-time and lifetime cumulative distribution functions. Error bars reflect the standard errors of the mean for three replicates of an experiment where the eIF4E–mRNA binding rate is measured across at least 100 mRNA molecules. (F) eIF4E–mRNA dissociation rates from the experiments in E. (G) eIF4E–mRNA equilibrium dissociation constants computed from the rates shown in E and F. (H) Dependence of eIF4E–mRNA association rate on mRNA length. The rates for JJJ1, HXT2, HSP30, NCE102, and MIM1 were measured in the present study. The rates for GIC1, SSA1 and ATP4, previously published in Çetin et al. (53), were added to include data points for correlation over a sufficient length range. (I) Dependence of eIF4E–mRNA association rate on coding-sequence length.
