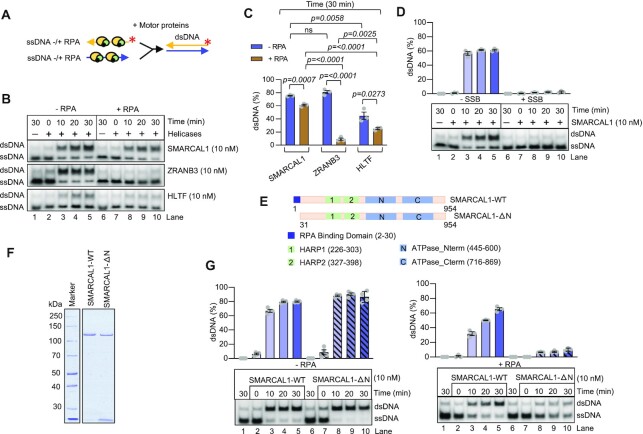
Figure 2.
SMARCAL1 anneals RPA-coated ssDNA. (A) A schematic of ssDNA annealing assay. (B) Annealing of ssDNA by SMARCAL1, ZRANB3 and HLTF without or with RPA (4 nM). Representative experiments are shown. (C) Quantification of experiment as in (B) at 30 min (error bars indicate SEM of three replicates). Two-tailed unpaired t-test was performed to generate the P values. (D) Annealing of ssDNA by SMARCAL1 without or with human mitochondrial SSB (4 nM). Top, quantifications (error bars indicate SEM of three replicates); bottom, representative experiment. (E) Top, a schematic showing domain organization of SMARCAL1. RPA binding domain is located in the N-terminal part of SMARCAL1 (indicated in dark blue). SMARCAL1-ΔN lacking RPA binding domain is shown below. (F) Recombinant SMARCAL1-WT and SMARCAL1-ΔN were analyzed by polyacrylamide gel electrophoresis and stained with Coomassie Brilliant Blue. (G) A comparison of SMARCAL1-WT and SMARCAL1-ΔN in ssDNA annealing without or with RPA (4 nM). Top, quantifications (error bars indicate SEM of three replicates); bottom, representative experiments.