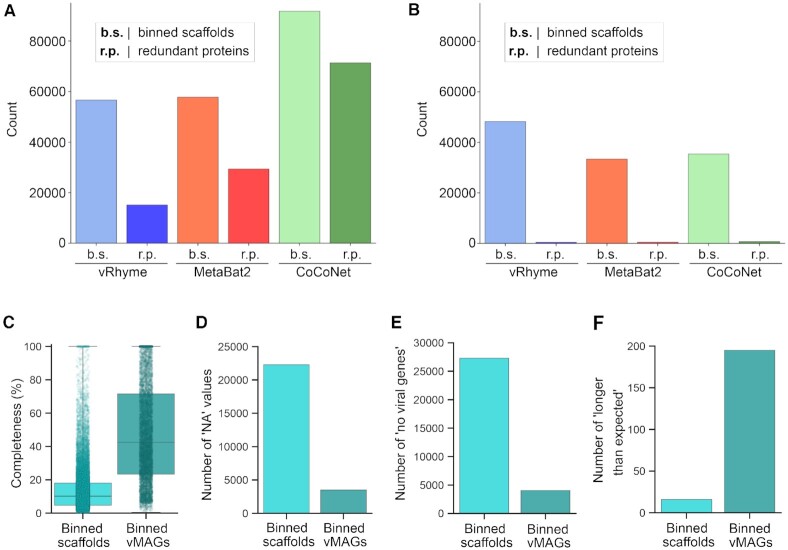
Figure 4.
Benchmark binning and genome completeness evaluation of GOV2. Comparison of vRhyme, MetaBat2, and CoCoNet (A) raw results and (B) low contamination filtering results by the number of scaffolds binned and identified redundancy. For vRhyme only, CheckV was used to identify (C) the estimated completeness values, (D) number of ‘NA’ completeness values, (E) number of ‘no viral genes’ scaffolds/vMAGs and (F) number of ‘longer than expected’ scaffolds/vMAGs for the low contamination results of individual binned scaffolds as well as vMAGs.