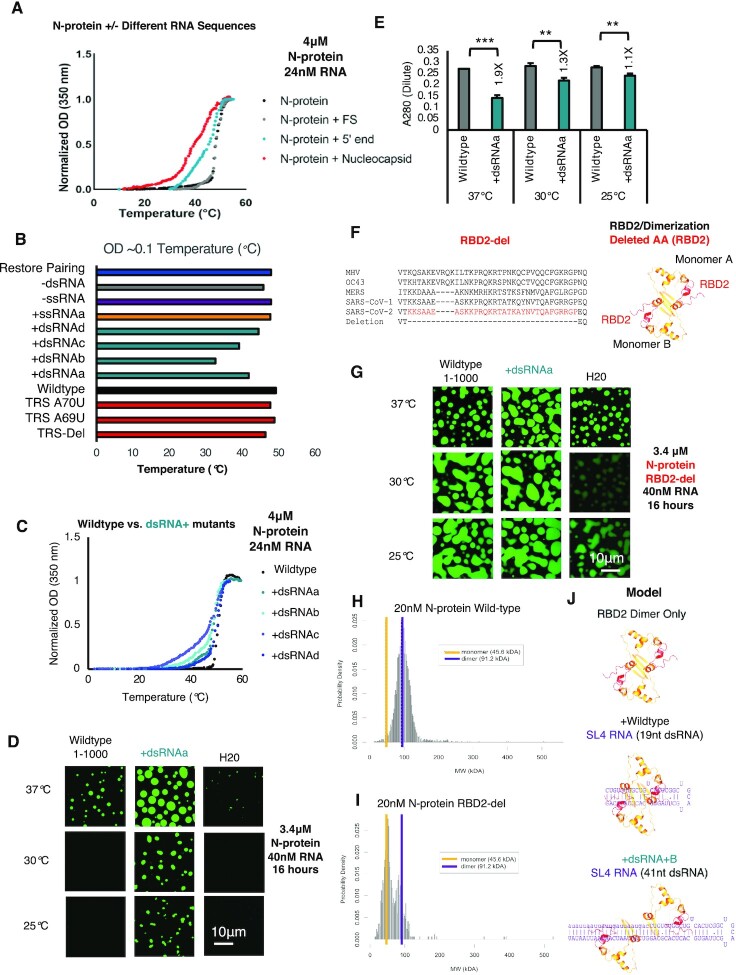
Figure 2.
RNA sequence and structure encodes N-protein LCST behavior via RBD2. (A) Temperature dependent turbidity tests of N-protein alone (Black), N-protein with Frameshifting region RNA (FS) (Gray), and N-protein with 5′end RNA (1–1000nt) (Blue) and N-protein with Nucleocapsid RNA (Red). Addition of droplet forming RNAs, 5′end 1–1000nt and Nucleocapsid RNA to N-protein, lowers the transition temperature but solubilizing RNA (FS) does not. (B) Transition temperature comparison (repeat of the experiment shown in (A) of wildtype 5′end or 11 mutants in the context of 1–1000nt. Bar length indicates the temperature in°Celsius at which the turbidity of the solution reaches ∼0.1. Only those mutants which alter the dsRNA content (teal + dsRNA), lower the temperature at which OD reaches ∼0.1 indicative of increased solution turbidity. (C) Temperature dependent turbidity tests for N-protein plus wildtype 5′end RNA as well as the four more structured mutants (+dsRNA) which lower the transition temperature. (D) Validation of the turbidity assay using droplet imaging (Figure 2B and C). 3.4μM Wildtype N-protein was mixed with either 40nM of wildtype 5′end 1–1000 RNA, +dsRNAa (RBD1 independent Figure 1G) or water only added control (H20) and incubated at the indicated temperature 37, 30, or 25°C for a period of 20 hours prior to imaging. Consistent with previous results, +dsRNAa increases droplet size relative to wildtype at 37°C (Figure 1F) & induces condensation at lower temperatures. (E) A280 measurement of remaining N-protein in the dilute phase for (D). At all temperatures, +dsRNAa lowers A280 measurements relative to wildtype. Error bars mark standard deviation for the three replicates and * indicate significance students t test (*** P < 0.001, ** P < 0.01, *P < 0.05, ns not significant) with brackets showing comparison for the indicated statistical test. (F) Protein sequence conservation of N-protein RBD2 and structure model of the RBD2 dimerization domain for SARS-CoV-2 (red sequences/red ribbon) indicate the location of the deletion in the primary sequence tested in (G). (G) RBD2/Dimerization domain is required for proper N-protein LCST behavior at indicated temperature range. 3.4 μM of N-protein RBD-del (green) was mixed with 25nM of either wildtype 1–1000, +dsRNAa, or water only control and incubated at the indicated temperatures for 16 h. Droplet formation was observed in all conditions although RNA dependence was more evident at lower protein concentrations (Supplementary Figure S4G-H). (H, I) Mass photometry histograms showing the molecular weight (MW) distribution of detected particles for wild-type N-protein (H) or RBD-del N-protein (I). (H) Wildtype N-protein is a stable dimer in solution (250 mM NaCl pH 7.5 20 mM phosphate buffer 20 nM N-protein) but RBD2-del is mostly a monomer (I). (J) Model of N-protein RBD2/Dimerization domain interactions with dsRNA. Binding of the two RBD2s of the two monomers of N-protein to dsRNA facilitates dimerization dissociation with temperature facilitating dissociation for shorter stem-loops. For all images scale bar indicates 10 μm all experiments show representative images from at least 3 replicates.