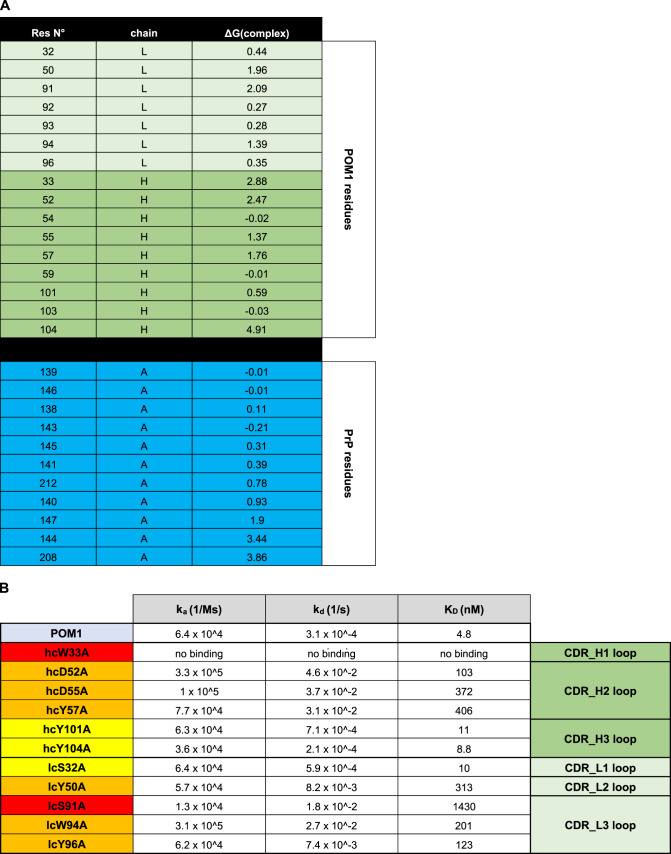
Table 1.
a, Computational alanine scanning indicates which residues of POM1 and PrP contribute to binding. Positive numbers in the third column suggest loss of binding energy. b, On the basis of these results (Table 1a), we prepared 11 single mutations of POM1 (in each CDR loop) as scFv constructs. Colors (yellow to red) visualize the impact on binding affinity. The mutated residues are shown as sticks on the cartoon POM1 structure in Extended Data Figure 5b