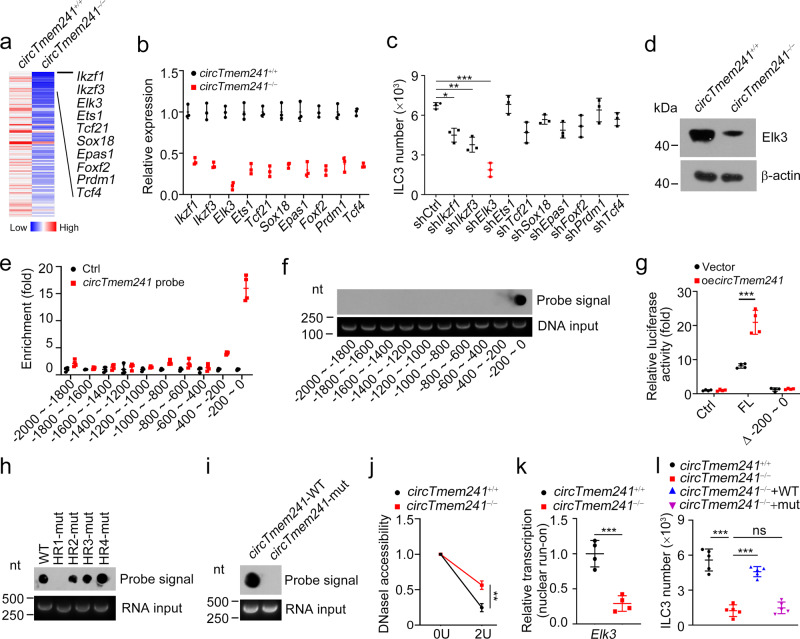
Fig. 4. CircTmem241 enhances Elk3 expression.
a Heatmap of top ten downregulated TFs in circTmem241−/− ILCPs versus circTmem241+/+ ILCPs by transcriptome analysis. b Relative expression levels of top 10 TFs in circTmem241+/+ and circTmem241−/− ILCPs were analyzed by qRT-PCR. Fold changes were normalized to endogenous Gapdh. n = 3 biological independent experiments. c Top 10 downregulated TFs were knocked down in ILCPs were analyzed by FACS. n = 3 biological independent experiments. d Elk3 protein levels in circTmem241+/+ or circTmem241−/− ILCPs by western blotting. e Enrichment of circTmem241 on Elk3 gene promoter was analyzed by CHIRP assay. CircTmem241 probe was biotin labeled. n = 4 for each group. f Enrichment of circTmem241 on Elk3 gene promoter was further validated by dot blot. g Luciferase reporter assay was performed to validate circTmem241 function on Elk3 transcription activation. FL, full-length (represent −2000~0 region upstream Elk3 transcriptional start site). n = 4 for each group. h, i CircTmem241 hairpin region (HR) sequence mutations were incubated with Elk3 promoter regions, followed by dot blot. j DNase I assay of chromatin accessibility in Elk3 promoters of circTmem241+/+ or circTmem241−/− ILCPs by qRT-PCR. n = 3 for each group. k Transcription activities of Elk3 in circTmem241+/+ or circTmem241−/− ILCPs were measured by nuclear run-on assay. n = 4 for each group. l ILCPs from circTmem241+/+ or circTmem241−/− mice were infected with virus expressing circTmem241-WT or circTmem241-mut, followed by in vitro differentiation assay. ILC3s (CD3−CD19−CD127+CD45loRORγt+) were analyzed by FACS. n = 5 for each group. WT, wild-type circTmem241; mut, circTmem241 mutation. *P < 0.05, **P < 0.01, and ***P < 0.001. Data were analyzed by an unpaired two-side Student’s t test and shown as means ± SD. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.