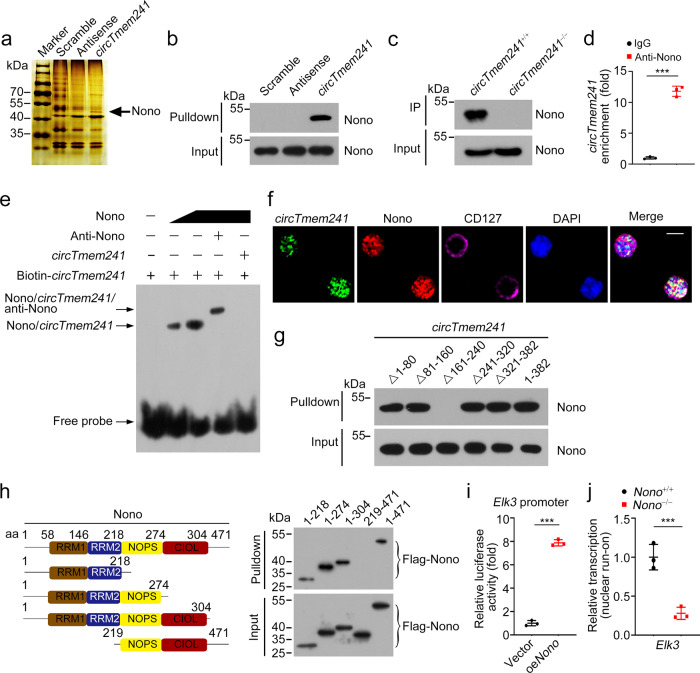
Fig. 5. CircTmem241 directly interacts with Nono.
a BM cells from WT mice were lysed and incubated with in vitro transcribed linear biotin-labeled circTmem241 transcripts, antisense, or scramble control. Pulldown components were separated by SDS-PAGE and followed by silver staining. Binding proteins of circTmem241 were identified via mass spectrometry. b BM cell lysates were incubated with circTmem241 probes or controls, followed by Western blotting. c Association between circTmem241 and Nono was validated by CHIRP assay, followed by immunoblotting. d RNA immunoprecipitation (RIP) assay was conducted using anti-Nono antibody or IgG through BM cell lysates and circTmem241 enrichment was detected by qRT-PCR. n = 3 for each group. e EMSA was performed using biotin-labeled circTmem241 transcripts and recombinant Nono proteins with or without anti-Nono antibody. f CircTmem241 was colocalized with Nono in ILCPs by immunofluorescence staining and confocal imaging. Scale bar, 10 μm. g RNA-pulldown assay was performed using indicated truncations of linear biotin-labeled circTmem241 RNAs with BM cell lysates. Interaction of Nono with indicated circTmem241 mutants was analyzed by western blotting. h Validation of binding ability of circTmem241 with indicated Nono truncations by RNA-pulldown assay, followed by western blotting. i Luciferase reporter assay was conducted to validate Nono function on Elk3 transcription activation. n = 3 for each group. j Transcription activity of Elk3 in Nono+/+ or Nono−/− ILCPs was measured by nuclear run-on assay. n = 3 for each group. ***P < 0.001. Data were analyzed by an unpaired two-side Student’s t test and shown as means ± SD. Data are representative of at least three independent experiments. Source data are provided as a Source Data file.