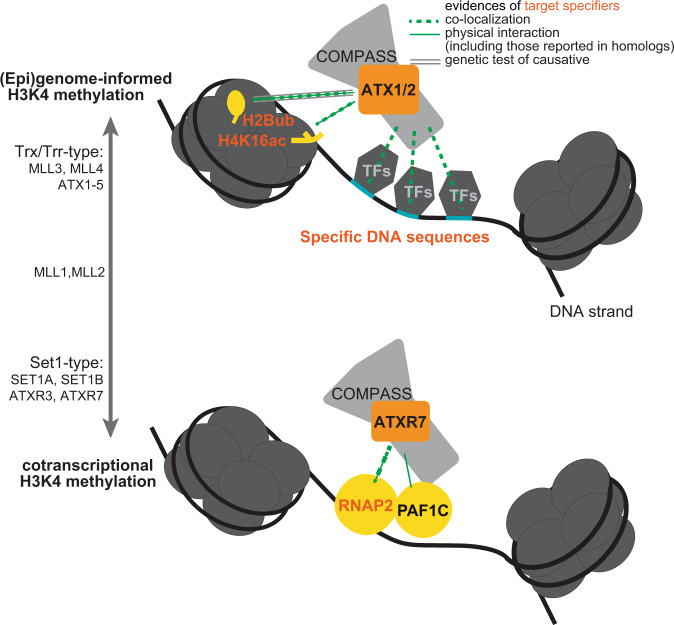
Fig. 8. Suggested models of (epi)genome-informed and cotranscriptional modes of H3K4 methylation.
Random forest and SVM modelings suggest that ATX1 and ATX2 localize based on chromatin features such as H2Bub, H4K16ac (Fig. 3) and several DNA sequences (Fig. 4). Mechanistic studies on ATX homologs indicate that H2Bub53 and H4K16ac52 tether H3K4 methyltransferases to nucleosomes. Some TFs may also bridge ATX to specific DNA sequences. On the other hand, ATXR7 is suggested to be cotranscriptional, based on its colocalization with RNAP2 (Fig. 3) and its contribution to the transcription-H3K4me1 correlation (Fig. 5). Studies in yeast homolog SET1 clarified Polymerase II Associated Factor 1 Complex (PAF1C) interacts with RNAP2 and COMPASS, thus providing a scaffold for H3K4 methyltransferases to work with RNAP26,7. These (epi)genome-informed and cotranscriptional modes of H3K4 methylation may also be generalized to other Arabidopsis H3K4 methyltransferases (Fig. 6) and other organisms (Fig. 7).