Figure 1.
Establishment of a COL7A1 mutation database specific for Korean RDEB patients and analysis of Korean and worldwide databases
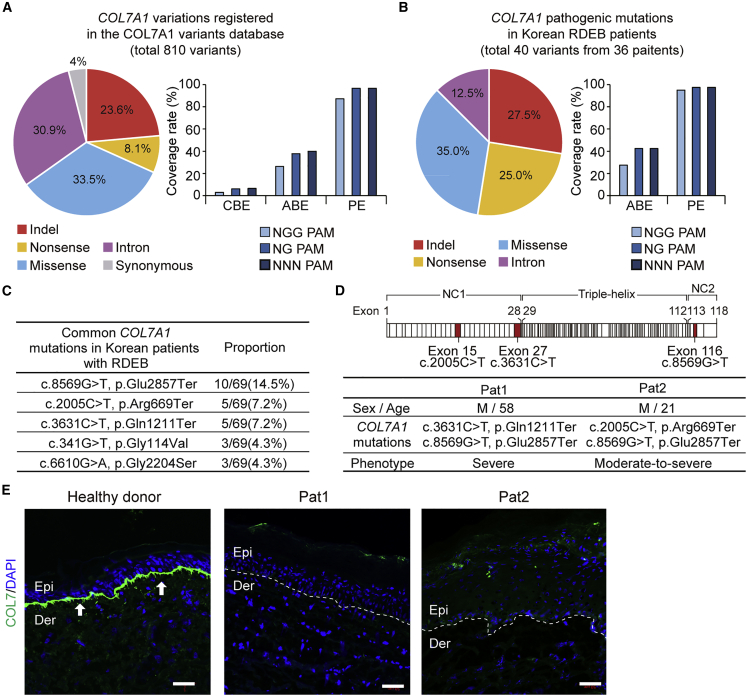
(A) The full spectrum of pathogenic COL7A1 mutations reported to date in DEB patients. Frequencies (%) of COL7A1 alleles that can theoretically be corrected with CBE, ABE, and PE constructed with SpCas9 (NGG-PAM) or its variant with a relaxed PAM requirement (NG-PAM) are shown. (B) Mutational analysis of COL7A1 in a large cohort of Korean RDEB patients. Frequencies (%) of COL7A1 alleles that can theoretically be corrected with ABE and PE recognizing NGG- or NG-PAMs are shown. (C) The five most frequent pathogenic COL7A1 mutations in our database among the 36 Korean RDEB patients with 69 pathogenic mutations. (D) Information about the two RDEB patients enrolled in this study and a schematic representation of procollagen VII showing the locations of the COL7A1 mutations identified in these patients. (E) Immunofluorescence to visualize C7 was performed on skin samples from Pat1, Pat2, and healthy controls using polyclonal rabbit anti-COL7 antibody. The white dotted line indicates the DEJ. White arrows indicate C7 deposited at the DEJ. Representative images are shown. Scale bars, 50 μm. Epi, epidermis; Der, dermis.