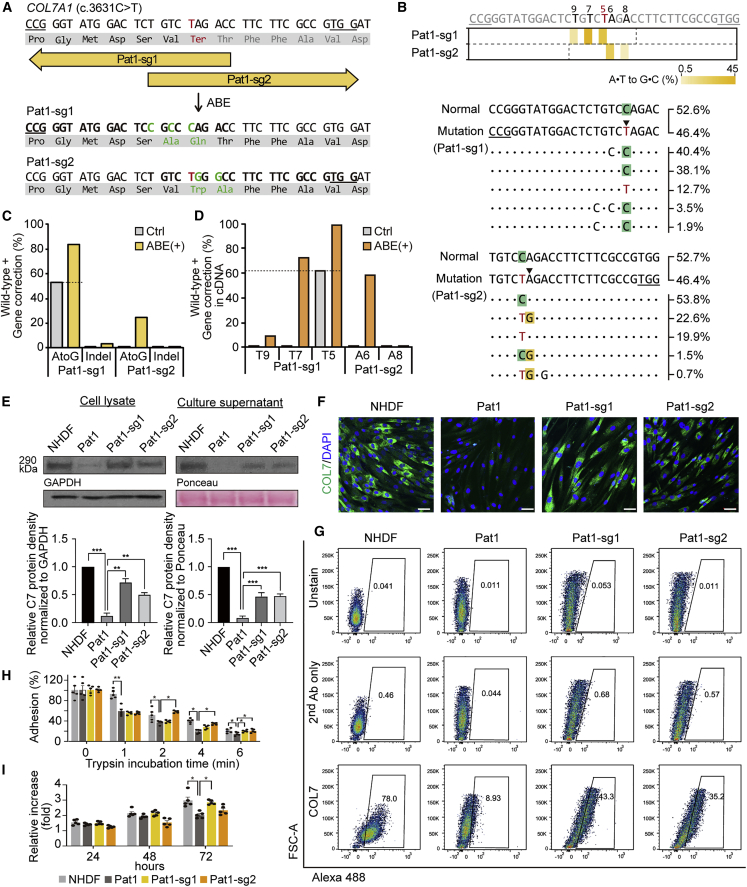
Figure 2.
Correction of c.3631C > T (Q1211X) in COL7A1 using ABEs in primary RDEB fibroblasts
(A) Schematic diagram of the ABE target sequence in exon 27 of the COL7A1 gene containing a C > T nonsense mutation (c.3631C > T, p.Q1211X). The mutant sequence is shown in red. Target sequences are highlighted in bold type, and PAM sequences are underlined. A·T to G·C conversions in the ABE editing window are shown in green. (B) Heatmap visualizing A·T to G·C conversion rates analyzed by high-throughput sequencing (top) and the sequences at the target sites together with their proportions (middle and bottom). The five most common sequences in ABE-treated RDEB fibroblasts are shown, and the frequencies of normal and mutated alleles in the non-edited patient-derived fibroblasts are shown at the top. PAM sequences are underlined, and the target A·T is indicated by an arrowhead. (C and D) Conversion rates calculated by deep sequencing of genomic DNA (C) and mRNA (D) from RDEB fibroblasts treated with different sgRNAs. (E) Western blots to measure C7 abundance in cell lysates and culture supernatants using GAPDH and Ponceau as internal controls, respectively. Images are representative of three independent experiments. Protein band densities from three independent experiments are presented as bar graphs. Each density value was normalized to the loading control value and expressed relative to the value in NHDFs. Data are the mean ± SEM (n = 3); ∗∗p < 0.01, ∗∗∗p < 0.001. (F) Immunofluorescence staining to visualize the C7 protein (green) in NHDFs, non-edited RDEB fibroblasts from Pat1, and ABE-treated RDEB fibroblasts. Nuclei were stained with DAPI (blue). Shown are representative images from at least three independent cell samples. Scale bars, 50 μm. (G) Representative graphics of flow cytometry assay for C7 restoration and quantitation of the percentage of C7-positive cells (n = 2). Histograms for unstained control and secondary antibody control for each cell samples are also shown. (H) Trypsin-based cell detachment assay. Cell adhesion is represented as the percentage of cells that remain attached after the indicated period of trypsin treatment. Five independent experiments were performed. Data are the mean ± SEM; ∗p < 0.05, ∗∗p < 0.01. (I) The proliferation of NHDFs, non-edited fibroblasts from Pat1, and ABE-treated RDEB fibroblasts was evaluated by the WST-1 assay. The ratio of the absorbance at 24, 48, and 72 h to that at 0 h is shown. Five independent experiments were performed. Data are the mean ± SEM; ∗p < 0.05. Statistical analyses were performed using unpaired Student’s t test for (E), (H), and (I) (NHDF versus Pat1 group) and one-way ANOVA followed by Dunnett’s tests for (E), (H), and (I) (each group compared with Pat1).