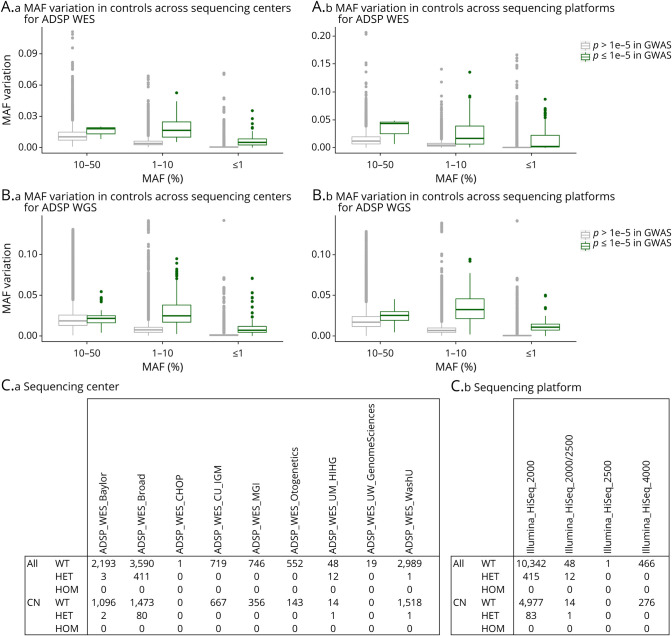
Figure 1. Variant Artifacts Across Different Sequencing Centers/Platforms Drive Spurious Associations in ADSP WES and WGS data.
In initial exome-wide and genome-wide association studies of ADSP WES and WGS, we observed many spurious associations (p ≤ 1e−5) using model 1 (i.e., not adjusting for sequencing center/platform; cf. Figures 2A and 3A). On inspection of these signals, it was notable that these variants displayed large variation in genotype counts across sequencing centers/platforms. The MAF variation in controls for all analyzed variants is visualized in (A.a-b) for ADSP WES and in (B.a-b) for ADSP WGS. (C.a-b) A specific example of a variant showing spurious association is provided. This variant, rs199707443, has an MAF of 0.003% in non-Finnish Europeans in Genome Aggregation Database v3.1.1, contrasting the 411 heterozygote counts in the Broad sequencing center. Notably, this particular variant still showed genome-wide significant association with Alzheimer disease risk even after sequencing center/platform adjustment (cf. Figure 2B). ADSP, Alzheimer Disease Sequencing Project; CN, cognitively normal; HET, heterozygote; HOM, homozygote; MAF, minor allele frequency; WT, wild type; WES, whole-exome sequencing; WGS, whole-genome sequencing.