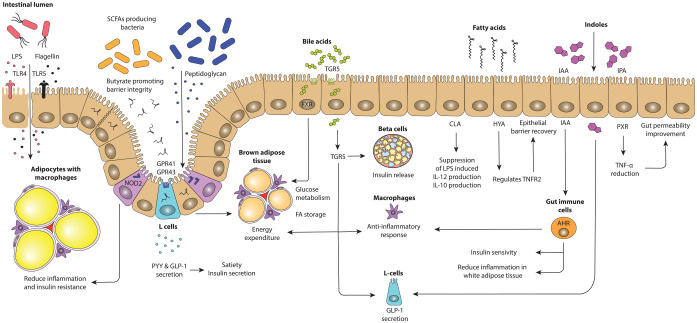
Figure 1.
Putative mechanisms by which the gut microbiota can influence host metabolism. Parts of the gut microbiota, such as flagellin, and LPS bind to TLRs,24,27 whereas intracellular NOD2 senses peptidoglycan. 28 Production of several SCFAs can bind to GPR41 and GPR43 leading to increased expression of PYY and GLP-1. 29 Bile acids activate TGR5 and FXR affecting lipid and glucose metabolism.30,31. Fatty acids, such as HYA, regulate TNFR2, involved in epithelial barrier recovery. 32 Indoles influence the host metabolism via GLP-1 modulation 33 and activation of AHR and binding to PXR.34,35
AHR, aryl hydrocarbon receptor; GLP-1, glucagon-like peptide-1; GPR, G protein-coupled receptor; HYA, 10-hydroxy-cis-12-octadecenoic acid; IAA, indole-3-acetic-acid; IPA, indole-3-propionic acid; LPS, lipopolysaccharide; NOD2, nucleotide-binding oligomerization domain 2; PXR, pregnane X receptor; PYY, peptide YY; SCFAs, short-chain fatty acids; TGR5, Takeda G protein-coupled receptor; TLRs, toll-like receptors; TNFR2, tumour necrosis factor receptor 2.