Abstract
Background
Plasma bedaquiline clearance is reportedly more rapid with African ancestry. Our objective was to determine whether genetic polymorphisms explained between-individual variability in plasma clearance of bedaquiline, its M2 metabolite, and clofazimine in a cohort of patients treated for drug-resistant tuberculosis in South Africa.
Methods
Plasma clearance was estimated with nonlinear mixed-effects modeling. Associations between pharmacogenetic polymorphisms, genome-wide polymorphisms, and variability in clearance were examined using linear regression models.
Results
Of 195 cohort participants, 140 were evaluable for genetic associations. Among 21 polymorphisms selected based on prior genome-wide significant associations with any drug, rs776746 (CYP3A5∗3) was associated with slower clearance of bedaquiline (P = .0017) but not M2 (P = .25). CYP3A5∗3 heterozygosity and homozygosity were associated with 15% and 30% slower bedaquiline clearance, respectively. The lowest P value for clofazimine clearance was with VKORC1 rs9923231 (P = .13). In genome-wide analyses, the lowest P values for clearance of bedaquiline and clofazimine were with RFX4 rs76345012 (P = 6.4 × 10−7) and CNTN5 rs75285763 (P = 2.9 × 10−8), respectively.
Conclusions
Among South Africans treated for drug-resistant tuberculosis, CYP3A5∗3 was associated with slower bedaquiline clearance. Different CYP3A5∗3 frequencies among populations may help explain the more rapid bedaquiline clearance reported in Africans. Associations with RFX4 and CNTN5 are likely by chance alone.
Keywords: bedaquiline, clofazimine, pharmacogenomics, tuberculosis, pharmacokinetics
In a cohort of patients treated for drug-resistant tuberculosis in South Africa, CYP3A5∗3was associated with slower plasma bedaquiline clearance. Different CYP3A5∗3minor allele frequencies among populations may help explain the more rapid bedaquiline clearance previously reported with African ancestry.
Drug-resistant tuberculosis is a major public health threat, with the number of patients infected with Mycobacterium tuberculosis resistant to rifampicin and isoniazid (ie, multidrug-resistant [MDR] tuberculosis) increasing worldwide. Treatment of MDR tuberculosis requires 9–24 months of therapy, and outcomes are less favorable than with drug-susceptible tuberculosis. Outcomes can be improved with novel and repurposed drugs. Clinical trials are evaluating 6-month regimens to simplify management and improve outcomes [1].
Bedaquiline is an oral agent active against M. tuberculosis that is resistant to first- and second-line drugs [2–4]. In 2012, bedaquiline received regulatory approval for treating pulmonary MDR tuberculosis as part of combination therapy [5]. It is part of standard-of-care regimens for MDR tuberculosis [6], and it has been associated with reduced all-cause mortality rates [7]. Bedaquiline is well absorbed, its plasma exposure increases proportionally with increasing dose, and food increases its oral bioavailability [2, 3]. Bedaquiline is highly bound to plasma proteins and has a long terminal elimination half-life, likely reflecting slow release from peripheral tissues. Model-based analyses describing exposure-response relationships predict that higher bedaquiline exposure would improve mycobacterial treatment responses [8, 9].
Bedaquiline is primarily metabolized by hepatic cytochrome P450 (CYP) 3A4 into an N-monodesmethyl metabolite (M2), which is approximately 5-fold less active against M. tuberculosis than bedaquiline [10]. A minor N-didesmethyl metabolite (M3) lacks antimycobacterial activity. Both M2 and M3 are more toxic in vitro than is bedaquiline, based on cytotoxicity assays and induction of phospholipidosis [10, 11]. Bedaquiline-associated QT prolongation may also depend on M2 exposure [12]. Although CYP isoforms 1A1, 2C8 and 2C18 metabolize bedaquiline in vitro at rates ≥10% that of CYP3A4, they are unlikely to contribute substantially in vivo, given their considerably lower expression levels in liver compared with CYP3A4 [13]. In a population pharmacokinetic model, apparent clearance of bedaquiline was 52% more rapid among individuals identified as being of black race compared with others [14], suggesting that genetic polymorphisms may affect disposition. A subsequent population pharmacokinetic analysis showed that weight and albumin levels, which were inversely correlated with each other, were significantly associated with bedaquiline plasma disposition. Age and race were also significant covariates [15]. While generally well tolerated, bedaquiline has been associated with QT prolongation, arthralgias, headache, and hepatic transaminase elevation.
Clofazimine is an oral lipophilic riminophenazine antibiotic discovered in 1957. Although historically used to treat leprosy, it is now recommended for treating rifampicin-resistant tuberculosis [16, 17]. Its oral bioavailability is approximately 70% [18], and administration with a high-fat meal increases its area under the concentration-time curve by 60% [19]. Clofazimine is highly protein bound [20] and undergoes duration-dependent accumulation in fat, macrophages, and reticuloendothelial organs, resulting in a very long terminal elimination half-life [21]. It is metabolized by hydrolytic reactions and is excreted largely unchanged [22, 23]. Adverse reactions to clofazimine include skin discoloration and QT prolongation. The present analyses leveraged population pharmacokinetic modeling data from patients treated for drug-resistant pulmonary tuberculosis in South Africa to characterize associations between genetic polymorphisms and between-individual variability in plasma clearance of bedaquiline, its M2 metabolite, and clofazimine.
METHODS
Study Population
The Pharmacokinetics, Resistance, and Outcomes of Bedaquiline in MDR- and XDR-TB (PROBeX) study was a prospective observational cohort project conducted between 2016 and 2020 at 3 tuberculosis referral hospitals in the South African provinces of Eastern Cape, Western Cape, and KwaZulu-Natal [24]. Participants received a modified standardized regimen, which typically included bedaquiline (400 mg once daily for 2 weeks, followed by 200 mg 3 times weekly), clofazimine (100 mg once daily), linezolid (600 mg daily), levofloxacin (750–1000 mg daily), ethionamide (15–20 mg/kg; maximum 750 mg daily), terizidone (15–20 mg/kg; maximum 750 mg daily), and pyrazinamide (20–30 mg/kg; maximum 1600 mg daily). All human immunodeficiency virus (HIV)–positive participants were offered either nevirapine- or lopinavir-ritonavir-based antiretroviral therapy to avoid CYP3A4 induction by efavirenz [25].
Pharmacokinetic Parameters
In PROBeX, clinical and laboratory data were collected monthly for 6 months, and every 6 months thereafter. Sparse pharmacokinetic sampling was performed at approximately 1, 2, and 6 months after the start of treatment, at single time points after self-reported dosing. A subgroup of consecutive participants underwent intensive sampling at month 2 (before and 1, 2, 3, 4, 5, 6, 8, and 24 hours after an observed dose and standard meal). Bedaquiline and clofazimine plasma concentrations were measured at the Division of Clinical Pharmacology of the University of Cape Town, using validated liquid chromatography with tandem mass spectrometry assays with interday accuracy ranging from 101% to 105% and precision (percentage coefficient of variation [%CV]) ranging from 3.3% to 4.6% during sample analysis for clofazimine. The accuracy statistics of the low-, medium-, and high-quality control samples of both bedaquiline and M2 during sample analysis were between 95.1% and 100.1%, with precision (%CV) between 4.2%, and 7.7% [26, 27].
Population pharmacokinetics were described with nonlinear mixed-effects models, which comprise a structural component (fixed effects) and a stochastic component (random effects). The stochastic model divides unexplained variabilities into between-subject variability or within-subject variability assigned to specific parameters and the residual error.
A previous pharmacokinetic analysis of bedaquiline and M2 included data from patients with MDR tuberculosis from 2 phase IIb studies (NCT00449644 and NCT00910871). Bedaquiline and M2 disposition were well described by 3- and 1-compartment models, respectively. Weight and albumin were correlated, typically increased after starting treatment, and significantly affected bedaquiline and M2 plasma disposition. Age and race were also significant covariates [15]. Because race is related to genetics, we prevented the model from explaining variability using race by fixing the race effect to zero and weighing the clearances of the black and nonblack groups to determine the typical clearance for the full population. In addition, we incorporated the known effect of concomitant lopinavir-ritonavir treatment on bedaquiline and M2 clearances by fixing the effect sizes to previously reported values [28]. The model was then fitted to PROBeX participants with maximum a posteriori estimation.
A population pharmacokinetic model for clofazimine was reported elsewhere [26]. The population model was developed based on pooled data from a phase 2A 14-day early bactericidal activity trial of clofazimine, alone or in combination with other antituberculous drugs (NCT 01691534) [29], and from the PROBeX study [26]. Based on data from 139 participants, clofazimine pharmacokinetics were well characterized by a 3-compartment model. Body composition was found to be a key covariate affecting drug exposures and disposition.
Genetic Polymorphisms
Whole-blood samples collected from consenting participants were labeled with coded identifiers. DNA was extracted at the Centre for Proteomic and Genomic Research in Cape Town, by a method described elsewhere [30]. Genotyping was performed using the Illumina Infinium Multi-Ethnic Global BeadChip array (MEGAEX), and postgenotyping quality control was performed by Vanderbilt Technologies for Advanced Genomics (VANTAGE). Quality control steps were performed using PLINK software, version 1.9 [31]. Genotyping efficiency per participant was 95% for all samples. To identify polymorphisms not directly genotyped, data were then imputed using the TOPMed program [32] after transforming to genome build 38 using liftOver software (version 3.33) [33]. Imputed polymorphisms were excluded if they had imputation scores <0.3, genotyping call rates <99%, minor allele frequency <0.05, or Hardy-Weinberg equilibrium P values <1.0 × 10−8.
To adjust for genetic ancestry, we estimated continuous axes of ancestry incorporating the intersection of common autosomal genotypes using the EIGENSTRAT software package (version 6.0.1) [34]. We also included the 1000 Genomes Project phase 3 to provide global reference populations [35]. Principal components scree plots were inspected to ensure the components selected for analyses represented ancestral information; based on these plots, 2 principal components sufficiently adjusted for ancestry. Additional covariates were not included in association analyses because these were already evaluated and included as appropriate in population pharmacokinetic models. The bedaquiline model considered the weighted average of the typical value for nonblack and the typical value for black participants, with weighting based on the proportion of black relative to nonblack participants in the data set in which the model was developed. The bedaquiline model also accounted for difference in exposure in patients receiving lopinavir-ritonavir, which increases bedaquiline exposure [28]. The clofazimine model accounted for the effect of body composition on disposition parameters.
There was not a strong a priori rationale to focus on specific genes or polymorphisms, other than the CYP3A locus for bedaquiline. To decrease the burden of multiple testing, we followed an approach that stepwise prioritized sets of polymorphisms to interrogate. We reasoned that polymorphisms previously associated with ≥1 drug-related phenotype, or previously significantly associated with any trait genome wide, are more likely to be true associations than are polymorphisms not previously associated with any drug or trait. We used as references the Pharmacogenomics Knowledgebase (PharmGKB) [36] and the NHGRI-EBI (National Human Genome Research Institute–European Bioinformatics Institute) GWAS Catalog [37]. In PharmGKB, 173 polymorphisms were previously associated with ≥1 drug-related phenotype (pharmacokinetics, efficacy, or toxicity), with levels of evidence of 1 (the preponderance of evidence shows an association, replicated in multiple cohorts and preferably with strong effect size) or 2 (moderate evidence of association; replicated, but some studies may not show statistical significance or may show small effect size). In the GWAS Catalog, 89 716 polymorphisms were previously associated with any trait at P < 5.0 × 10−8 in ≥1 published study. A subset of 33 polymorphisms were common to both PharmGKB and the GWAS Catalog. A list of PharmGKB and GWAS Catalog polymorphisms included in our analyses are provided in the Supplementary Materials.
We considered polymorphisms common to both PharmGKB and the GWAS Catalog to have the strongest a priori evidence for true associations. We secondarily evaluated all polymorphisms from PharmGKB and from the GWAS Catalog (based on criteria described above) and all polymorphisms in our imputed genome-wide data.
Association Analyses
Outcomes of primary interest were between-individual variability in population parameter estimates for central clearance of bedaquiline and clofazimine. We also studied between-individual variability in clearance of the M2 metabolite of bedaquiline. Multivariable linear regression models were used to characterize associations with genetic polymorphisms, using 2-sided statistical tests. The first 2 genetic ancestry principal components were included as covariates. We report the regression coefficient (β) for additive associations with polymorphisms, where positive β values indicate positive associations. To correct for multiple testing, the Bonferroni method was used to determine significance thresholds, with .05 divided by the number of polymorphisms tested in prioritized analyses, and P = 5.0 × 10 − 8 for genome-wide analyses. Linkage disequilibrium (LD) estimates were determined within our data set using PLINK software. We used LocusZoom software (version 0.12.0) to visualize genetic associations and LD estimates in defined regions [38].
Ethical Approval
The PROBeX study was approved by the institutional review boards at the University of Cape Town, Albert Einstein College of Medicine, and Emory University. All participants provided written informed consent.
RESULTS
Participant Characteristics
A total of 195 individuals were enrolled in the PROBeX cohort. All were ≥18 years of age and had baseline creatinine and alanine aminotransferase values no more than 2 or 5 times the upper limit of normal, respectively. Among participants, 123 (63%) were HIV positive, the median age was 33 years, 160 (82%) were black, and 111 (57%) were female. During the study period, 190 (97%) received clofazimine, and 179 (92%) received linezolid. Of the HIV-positive participants, 113 (90%) were receiving antiretroviral therapy before enrollment, and 26 (23%) received lopinavir-ritonavir during the study.
Among cohort participants, 172 and 164 provided population pharmacokinetic data for bedaquiline and clofazimine, respectively, of whom 140 and 136, respectively, were included in genetic association analyses. The primary reason for exclusion was genotyping efficiency <95%. Individuals included in genetic analyses closely resembled the total PROBeX cohort (Table 1).
Table 1.
Baseline Characteristics of Study Participants
| Characteristic | Total PROBeX Cohort (n = 195) | Bedaquiline Genetic Analysis Group (n = 140) | Clofazimine Genetic Analysis Group (n = 136) |
|---|---|---|---|
| Age, median (IQR), y | 33 (28−42) | 33 (27–41) | 33 (27–42) |
| Female sex, no. (%) | 111 (57) | 71 (51) | 71 (51) |
| Race, no. (%) | |||
| Black | 160 (82) | 112 (80) | 106 (78) |
| Mixed race | 33 (17) | 26 (19) | 28 (21) |
| White | 2 (1) | 2 (1) | 2 (1) |
| BMI, median (IQR)a | 20 (18–23) | 20 (18–23) | 20 (18–23) |
| HIV positive, no. (%) | 123 (63) | 82 (59) | 80 (59) |
Abbreviations: BMI, body mass index; HIV, human immunodeficiency virus; IQR, interquartile range; PROBeX, Pharmacokinetics, Resistance, and Outcomes of Bedaquiline in MDR- and XDR-TB.
BMI calculated as weight in kilograms divided by height in meters squared.
Associations With Between-Individual Variability in Bedaquiline and M2 Clearance
We primarily characterized associations with between-individual variability in plasma bedaquiline clearance. Of 33 polymorphisms common to PharmGKB and the GWAS Catalog (described in Methods), we were able to test for associations with 21 (64%). The other 12 polymorphisms failed imputation score, minor allele frequency, or Hardy-Weinberg equilibrium cutoffs. For bedaquiline clearance, the lowest P value among these 21 polymorphisms was for CYP3A5 rs776746 (P = .0017; β = −.18), with the C allele associated with slower clearance. This withstood correction for multiple testing (cutoff, 0.0024). This T→C polymorphism defines the CYP3A5∗3 allele which results in nonfunctional CYP3A5 protein [39, 40].
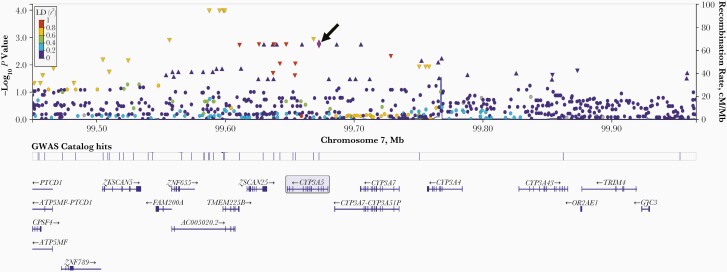
Given the association of CYP3A5∗3 with bedaquiline clearance, we more thoroughly interrogated the CYP3A gene locus, including 150 kB on either side of CYP3A4 and CYP3A5. Among 1214 polymorphisms in this region, the lowest P values for association with bedaquiline clearance were seen with 4 polymorphisms, approximately 73 kB 3’ of rs776746 (rs1011024, rs10254729, rs12333760, and rs34777615) (each P = 1.0 × 10−4; β = −.25), which were in strong LD with CYP3A5 rs776746 (r2 = 0.70). Two additional polymorphisms were in complete LD with rs776746, rs6465750, and rs4646457 (r2 = 1.0). A LocusZoom plot for the CYP3A locus ±150 kB is presented in Figure 1.
Figure 1.
LocusZoom plot of associations with between-individual variability in bedaquiline clearance, across the CYP3A locus on chromosome 7. Represented are −log10P values among 140 individuals for 1214 polymorphisms in the region, ranging from −150 kB from the 3’ end of CYP3A5 to +150 kB from the 5’ end of CYP3A4. Purple diamond and arrow identify CYP3A5 rs776746 (CYP3A5∗3 [arrow]), which is used as the reference for the linkage disequilibrium (LD) values in the graph. Each other marker is color coded to indicate r2 value categories for LD with rs776746, as shown in the legend. These LD values are based on the “ALL” LocusZoom setting for the reference population
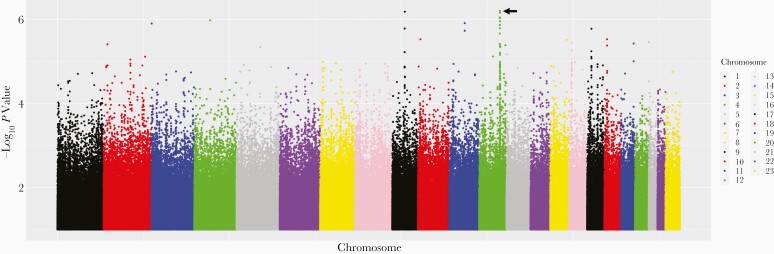
Secondarily considering PharmGKB polymorphisms that were not in the GWAS Catalog, we were able to test for associations with 36 of 136 (26%). The other 100 failed imputation score, minor allele frequency, or Hardy-Weinberg equilibrium cutoffs. For bedaquiline clearance, the lowest P value for association among these 36 polymorphisms was ERCC1 rs11615 (P = .081; β = .12). Considering polymorphisms previously associated with any GWAS Catalog trait, we were able to test for associations with 63 429 of 89 716 (71%). The lowest P value for association with bedaquiline clearance was RIC8B rs7977247 (P = 1.1 × 10−6; β = −.18). Considering genome-wide associations regardless of the GWAS Catalog, the lowest P value for association with bedaquiline clearance was for RFX4 rs763450 (P = 6.4 × 10−7; β = .20). A genome-wide Manhattan plot for bedaquiline clearance is presented in Figure 2. The 5 lowest P value polymorphisms for association with bedaquiline clearance in each of the above stepwise-prioritized analyses are provided in Table 2. Only CYP3A5 rs776746 withstood correction for multiple testing within any analysis. There were not strong genetic associations with between-individual variability in M2 clearance. For CYP3A5 rs776746 and M2 clearance, P = .25.
Figure 2.
Manhattan plot of associations with between-individual variability in bedaquiline clearance. Shown are associations among 140 individuals who received bedaquiline during participation in PROBeX and were evaluable for genetic associations. The lowest P value is for RFX4 rs763450 on chromosome 12 (arrow) (P = 6.4 × 10−7; β = .20).
Table 2.
Lowest P Values for Genetic Association With Between-Subject Variability in Plasma Bedaquiline Clearance in 140 PROBeX Participants
| Polymorphism | Gene | Chromosome | Reference Allele | Variant Allele | MAF | β Value | P Value |
|---|---|---|---|---|---|---|---|
| PharmGKB and GWAS Catalog (n = 21)a | |||||||
| rs776746 | CYP3A5 | 7 | T | C | 0.17 | −.18 | .0017 |
| rs8050894 | VKORC1 | 16 | C | G | 0.23 | .11 | .033 |
| rs1800629 | TNF | 6 | G | A | 0.14 | .11 | .068 |
| rs17782313 | MC4R | 18 | T | C | 0.24 | −.09 | .073 |
| rs12979860 | IFNL4 | 19 | T | C | 0.47 | .04 | .335 |
| PharmGKB, not GWAS Catalog (n = 36)a | |||||||
| rs11615 | ERCC1 | 19 | G | A | 0.13 | .12 | .08 |
| rs2359612 | VKORC1 | 16 | G | A | 0.26 | −.07 | .14 |
| rs7997012 | HTR2A | 13 | G | A | 0.05 | − .11 | .23 |
| rs1045642 | ABCB1 | 7 | G | A | 0.16 | −.07 | .24 |
| rs2298383 | ADORA2A | 22 | C | T | 0.38 | −.05 | .27 |
| GWAS Catalog (n = 63 502)a | |||||||
| rs7977247 | RIC8B | 12 | C | T | 0.43 | −.18 | 1.1 × 10−6 |
| rs10778495 | RFX4 | 12 | G | A | 0.47 | −.19 | 1.7 × 10−6 |
| rs10161520 | RFX4 | 12 | T | C | 0.49 | .17 | 1.2 × 10−5 |
| rs11113071 | RFX4 | 12 | T | C | 0.45 | .17 | 1.2 × 10−5 |
| rs10861637 | RFX4 | 12 | A | G | 0.28 | .19 | 1.3 × 10−5 |
| Genome-wide genotype datab (n = 9 074 402)a | |||||||
| rs763450 | RFX4 | 12 | A | G | 0.49 | .20 | 6.4 × 10−7 |
| rs78277930 | Intergenic | 9 | A | T | 0.05 | −.46 | 6.6 × 10−7 |
| rs80098193 | Intergenic | 9 | A | G | 0.05 | −.46 | 6.6 × 10−7 |
| rs114384536 | Intergenic | 9 | C | A | 0.05 | −.46 | 6.6 × 10−7 |
| rs12310706 | RFX4 | 12 | G | A | 0.45 | −.19 | 7.0 × 10−7 |
Abbreviations: MAF, minor allele frequency; PROBeX, Pharmacokinetics, Resistance, and Outcomes of Bedaquiline in MDR- and XDR-TB.
Parenthetical numbers are numbers of polymorphisms.
All polymorphisms with P values <1.0 × 10−6 are shown.
We next assessed whether including CYP3A5 rs776746 in the population pharmacokinetic model improved model fit. Based on log-likelihood profiling, rs776746 heterozygosity was associated with 15% slower (95% confidence interval, 3.5%–25.5%) and homozygosity with 30% slower (7.0%–51%) bedaquiline clearance. The model-predicted effects of rs776746 genotype on bedaquiline and M2 exposure are shown in the Supplementary Materials.
Genetic Associations With Between-Individual Variability in Clofazimine Clearance
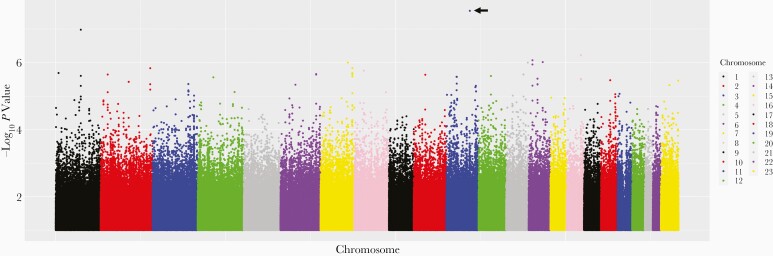
As we did for bedaquiline, we characterized associations with between-individual variability in plasma clofazimine clearance. We were able to test for associations with 21 of 33 polymorphisms (64%) common to PharmGKB and the GWAS Catalog. For clofazimine clearance, the lowest P value among these 21 polymorphisms was for VKORC1 rs9923231 (P = .13; β = .07). Considering polymorphisms in PharmGKB but not the GWAS Catalog, we were able to test for associations with 36 of 140 (26%). The lowest P value among these 36 polymorphisms was for IFNL3 rs11881222 (P = .047; β = .34). Considering polymorphisms previously associated with any trait in the GWAS Catalog, we were able to test for associations with 63 502 of 89 716 (71%). The lowest P value was for rs3827592, an intergenic chromosome 4 polymorphism (P = 1.7 × 10−5; β = .29). Considering genome-wide associations regardless of the GWAS Catalog, the lowest P value was for CNTN5 rs75285763 (P = 2.9 × 10−8; β = .46). A genome-wide Manhattan plot for clofazimine clearance is presented in Figure 3. The lowest P value polymorphisms for association with clofazimine clearance stratified by each of the above stepwise-prioritized analyses are presented in Table 3. Only CNTN5 rs75285763 was significant after correction for multiple testing within each analysis.
Figure 3.
Manhattan plot of associations with unexplained variability in clofazimine clearance. Shown are associations among 136 individuals who received clofazimine during participation in PROBeX and were evaluable for pharmacogenomics associations. The lowest P value is for CNTN5 rs75285763 on chromosome 11 (arrow) (P = 2.9 × 10−8; β = .46).
Table 3.
Lowest P Values for Genetic Association With Unexplained Variability in Plasma Clofazimine Clearance in 136 PROBeX Participants
| Polymorphism | Gene | Chromosome | Reference Allele | Variant Allele | MAF | β Value | P Value |
|---|---|---|---|---|---|---|---|
| PharmGKB and GWAS Catalog (n = 21)a | |||||||
| rs9923231 | VKORC1 | 16 | C | T | 0.07 | −.11 | .13 |
| rs887829 | UGT1A10 | 2 | C | T | 0.37 | −.05 | .17 |
| rs8099917 | Intergenic | 19 | T | G | 0.07 | .09 | .21 |
| rs489693 | Intergenic | 18 | A | C | 0.47 | −.04 | .23 |
| rs1800629 | TNF | 6 | G | A | 0.14 | .06 | .24 |
| PharmGKB, not GWAS Catalog (n = 36)a | |||||||
| rs11881222 | IFNL3 | 19 | A | G | 0.34 | .07 | .047 |
| rs2359612 | VKORC1 | 16 | G | A | 0.25 | −.07 | .058 |
| rs1042713 | ADRB2 | 5 | G | A | 0.42 | .06 | .083 |
| rs2740574 | LOC110366354 | 7 | C | T | 0.29 | −.06 | .115 |
| rs9934438 | VKORC1 | 16 | G | A | 0.07 | −.11 | .126 |
| GWAS Catalog (n = 63 502)a | |||||||
| rs3827592 | Intergenic | 4 | G | A | 0.29 | −.17 | 1.73 × 10−5 |
| rs7689452 | Intergenic | 4 | A | G | 0.29 | −.17 | 1.96 × 10−5 |
| rs79709502 | LINC01800 | 2 | C | G | 0.20 | .19 | 2.84 × 10−5 |
| rs5749446 | FBXO7 | 22 | T | C | 0.37 | −.16 | 3.21 × 10−5 |
| rs3827335 | FBXO7 | 22 | A | G | 0.38 | −.16 | 3.64 × 10−5 |
| Genome-wide genotype datab (n = 9 074 402)a | |||||||
| rs75285763 | CNTN5 | 11 | A | G | 0.04 | .46 | 2.87 × 10−8 |
| rs35274012 | Intergenic | 1 | G | A | 0.16 | .25 | 1.05 × 10−7 |
| rs562673502c | Intergenic | 16 | C | A | 0.05 | .39 | 6.05 × 10−7 |
| rs147293114 | NPAS3 | 14 | TTTAG | T | 0.05 | .40 | 8.47 × 10−7 |
| rs140444407 | Intergenic | 14 | C | T | 0.06 | −.35 | 9.62 × 10−7 |
| rs11769507 | Intergenic | 7 | C | T | 0.15 | .23 | 9.910 × 10−7 |
Abbreviations: MAF, minor allele frequency; PROBeX, Pharmacokinetics, Resistance, and Outcomes of Bedaquiline in MDR- and XDR-TB.
Parenthetical numbers are numbers of polymorphisms.
All polymorphisms with P < 1.0 × 10−6 are shown.
Polymorphism rs562673502 was in complete linkage with rs534375032, rs147681927, rs569881175, rs148413871, rs141244461, and rs141114616 in our imputed genotype data, so it has identical association results.
DISCUSSION
Bedaquiline and clofazimine are important for treating MDR tuberculosis. At the time of this writing, this is the first pharmacogenetic study of either bedaquiline or clofazimine. In our analyses that leveraged modeled pharmacokinetic data from the PROBeX cohort study of adults treated for MDR tuberculosis in South Africa, CYP3A5 rs776746 was associated with slower plasma bedaquiline clearance (P = .0017). When included as a covariate in the population pharmacokinetic model, rs776746 was significant, with heterozygosity associated with 15% slower and homozygosity with 30% slower clearance of bedaquiline. We also found a genome-wide significant association between CNTN5 rs75285763 and slower plasma clofazimine clearance (P = 2.9 × 10−8). Because both bedaquiline and clofazimine accumulate slowly in the body with repeated dosing, it was important that population pharmacokinetic models were used to evaluate their disposition.
Several considerations suggest that the association between CYP3A5 rs776746 and bedaquiline clearance is not by chance alone. First, bedaquiline is known to be metabolized by CYP3A4, and substrate specificity often overlaps between CYP3A4 and CYP3A5. The CYP3A5 rs776746 polymorphism (where T = CYP3A5∗1 and C = CYP3A5∗3) causes an alternatively spliced isoform that results in a premature stop codon and nonfunctional CYP3A5 protein [40]. Among individuals homozygous for CYP3A5∗3, CYP3A5 comprises only 5% of hepatic CYP3A expression, compared with as much as 50% among individuals carrying ≥1 copy of CYP3A5∗1 [39]. The CYP3A5∗3 allele also predicts plasma exposure of the immunosuppressant drug, tacrolimus [41]. Second, among 21 polymorphisms common to PharmGKB and the GWAS Catalog, and for which we characterized associations, CYP3A5 rs776746 had the lowest P value, was in the expected direction (ie, the C allele with slower bedaquiline clearance), and withstood correction for multiple testing. Finally, considering 1214 polymorphisms within the CYP3A locus, the polymorphisms with the lowest P value for association with bedaquiline clearance were in strong LD with CYP3A5 rs776746. Despite these considerations, the association between CYP3A5 rs776746 and bedaquiline clearance should be replicated in independent cohorts.
We found no association between CYP3A5 rs776746 and clearance of M2. As speculation, this could be explained by CYP3A isoforms being involved in both generating M2 from bedaquiline, and in metabolizing M2 to M3, which may make it harder to detect an effect on M2 clearance.
The CYP3A5 rs776746 loss-of-function C allele varies markedly in frequency depending on ancestry, ranging from approximately 30% among Africans to 70% among East Asians and 93% among Europeans [42]. Thus, Africans overall express substantially more CYP3A5 than do other populations. This may help to explain the 52% greater apparent clearance of bedaquiline among individuals identified as black in a previous population pharmacokinetic model [14]. In the present study the minor allele frequency of CYP3A5 rs776746 was 17%. Given that rs776746 is relatively frequent and that higher bedaquiline exposure may improve mycobacterial treatment responses [8, 9], genetic testing to guide bedaquiline dosing has the potential to improve treatment outcomes.
Regarding clofazimine clearance, we found a genome-wide significant association with CNTN5 rs75285763. We suspect that this is by chance alone. The gene CNTN5 encodes contactin 4, a member of the immunoglobulin superfamily [43]. Contactin 4 is a neuronal cell membrane adhesion molecule that helps to form axon connections in the developing nervous system. This protein seems unlikely to affect clofazimine exposure. In addition, this polymorphism is infrequent, with a minor allele frequency of only 0.05 in our study. Furthermore, no pharmacogenes reside near CNTN5 on chromosome 11, and the P value for rs75285763 (P = 2.9 × 10−8) was barely significant genome wide.
Other than the CYP3A locus with bedaquiline, there was no strong a priori evidence for an association of any specific polymorphisms or genes with bedaquiline or clofazimine pharmackinetics. For this reason, and to decrease the burden of multiple testing, we used an approach that leveraged the imputed genome-wide data generated with the PROBeX cohort against the vast knowledge generated by prior genetic association studies represented in PharmGKB [36] and the GWAS Catalog [37]. We reasoned that polymorphisms associated with ≥1 drug-related phenotype in PharmGKB with levels of evidence of 1 or 2 (as described in Methods), and also associated with any trait in the GWAS Catalog at P < 5.0 × 10−8 in ≥1 published study, would most likely be true-positives in the present study. This approach appeared to work well for CYP3A5 rs776746, which was common to PharmGKB and the GWAS Catalog. For secondarily prioritized analyses, we did not identify compelling associations (ie, in PharmGKB but not the GWAS Catalog, in the GWAS Catalog regardless of PharmGKB, and genome wide regardless of the GWAS Catalog),
The present study had limitations. The sample size was relatively modest. However, unlike genome-wide studies of some complex traits such as diabetes, large effect sizes with pharmacogenes and off-target genes often reveal significant associations with small sample sizes. The modest sample size limited our ability to detect associations with infrequent polymorphisms, those with small effect sizes, or those not previously associated with a drug or trait in PharmGKB or the GWAS Catalog. In addition to bedaquiline and clofazimine, cohort participants were receiving multiple medications to treat tuberculosis and HIV-1. Although not anticipated, it is conceivable that interactions between these drugs and bedaquiline or clofazimine may have obscured genetic associations. Finally, the present analyses focused on a cohort of African ancestry. Results may be different in other populations.
In summary, among patients treated for MDR tuberculosis in a prospective, observational cohort study in South Africa, the CYP3A5∗3 loss-of-function allele was associated with slower plasma clearance of bedaquiline. The variable frequency of this allele between populations suggests that CYP3A5∗3 may help to explain the more rapid clearance of bedaquiline reported among Africans. This association should be considered tentative until replicated in independent cohorts. A genome-wide significant association between a CNTN5 polymorphism and plasma clearance of clofazimine is likely by chance alone.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to all the participants in the PROBeX (Pharmacokinetics, Resistance, and Outcomes of Bedaquiline in MDR- and XDR-TB) cohort for making this study possible.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grants R01 AI114304, R01 AI145679, and K24 AI155045 to J. C. M. B. and K24 AI114444 to N. R. G.); Einstein-Rockefeller-CUNY (grant CFAR P30 AI124414); the Einstein/Montefiore Institute for Clinical and Translational Research (grant UL1 TR002556); the National Institutes of Health (grants AI069439, AI110527, AI077505, AI120790, and TR002243 to D. W. H. and K43 TW011421 to S. W.); This research was funded, in part, by the Wellcome Trust. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission (grants 098316, 214321/Z/18/Z, and 203135/Z/16/Z to G. Meintjes and 203135/Z/16/Z to S. W.); the South African Research Chairs Initiative of the Department of Science and Technology and the National Research Foundation of South Africa (grant 64787 to G. Meintjes); and the European & Developing Countries Clinical Trials Partnership (grant CDF1018 to S. W.).
Contributor Information
David W Haas, Department of Medicine, Vanderbilt University School of Medicine, Nashville, Tennessee, USA; Department of Internal Medicine, Meharry Medical College, Nashville, Tennessee, USA.
Mahmoud Tareq Abdelwahab, Division of Clinical Pharmacology, Department of Medicine, University of Cape Town, South Africa.
Stijn W van Beek, Department of Pharmacy, Radboud Institute for Health Sciences, Radboud University Medical Center, Nijmegen, the Netherlands.
Paxton Baker, Vanderbilt Technologies for Advanced Genomics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Gary Maartens, Division of Clinical Pharmacology, Department of Medicine, University of Cape Town, South Africa.
Yuki Bradford, Department of Genetics, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Marylyn D Ritchie, Department of Genetics and Institute for Biomedical Informatics, University of Pennsylvania Perelman School of Medicine, Philadelphia, Pennsylvania, USA.
Sean Wasserman, Division of Infectious Diseases, Department of Medicine, University of Cape Town, South Africa.
Graeme Meintjes, Wellcome Centre for Infectious Diseases Research in Africa, Institute of Infectious Disease and Molecular Medicine; Department of Medicine, University of Cape Town, Cape Town, South Africa.
Karen Beeri, Vanderbilt Technologies for Advanced Genomics, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Neel R Gandhi, Departments of Epidemiology & Global Health, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA; Division of Infectious Diseases, Department of Medicine, Emory School of Medicine, Emory University, Atlanta, Georgia, USA.
Elin M Svensson, Department of Pharmacy, Radboud Institute for Health Sciences, Radboud University Medical Center, Nijmegen, the Netherlands; Department of Pharmacy, Uppsala University, Uppsala, Sweden.
Paolo Denti, Division of Clinical Pharmacology, Department of Medicine, University of Cape Town, South Africa.
James C M Brust, Division of General Internal Medicine, Department of Medicine, Albert Einstein College of Medicine, Bronx, New York, USA.
References
- 1. Lange C, Dheda K, Chesov D, Mandalakas AM, Udwadia Z, Horsburgh CR Jr. Management of drug-resistant tuberculosis. Lancet 2019; 394:953–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andries K, Verhasselt P, Guillemont J, et al. . A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 2005; 307:223–7. [DOI] [PubMed] [Google Scholar]
- 3. Diacon AH, Pym A, Grobusch M, et al. . The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med 2009; 360:2397–405. [DOI] [PubMed] [Google Scholar]
- 4. Diacon AH, Donald PR, Pym A, et al. . Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother 2012; 56:3271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sirturo (bedaquiline) tablets prescribing information. Titusville, NJ: Janssen. https://www.janssenlabels.com/package-insert/product-monograph/prescribing-information/SIRTURO-pi.pdf. Accessed 25 February 2021. [Google Scholar]
- 6. World Health Organization. WHO consolidated guidelines on tuberculosis, module 4: treatment—drug-resistant tuberculosis treatment. https://www.who.int/publications/i/item/9789240007048. Accessed 9 December 2021. [PubMed]
- 7. Wang MG, Wu SQ, He JQ.. Efficacy of bedaquiline in the treatment of drug-resistant tuberculosis: a systematic review and meta-analysis. BMC Infect Dis 2021; 21:970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Svensson EM, Karlsson MO.. Modelling of mycobacterial load reveals bedaquiline’s exposure-response relationship in patients with drug-resistant TB. J Antimicrob Chemother 2017; 72:3398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanneau L, Karlsson MO, Svensson EM.. Understanding the drug exposure-response relationship of bedaquiline to predict efficacy for novel dosing regimens in the treatment of multidrug-resistant tuberculosis. Br J Clin Pharmacol 2020; 86:913–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Heeswijk RP, Dannemann B, Hoetelmans RM.. Bedaquiline: a review of human pharmacokinetics and drug-drug interactions. J Antimicrob Chemother 2014; 69:2310–8. [DOI] [PubMed] [Google Scholar]
- 11. Mesens N, Steemans M, Hansen E, Verheyen GR, Goethem F V, Gompel J V.. Screening for phospholipidosis induced by central nervous drugs: comparing the predictivity of an in vitro assay to high throughput in silico assays. Toxicol In Vitro 2010; 24:1417–25. [DOI] [PubMed] [Google Scholar]
- 12. Tanneau L, Svensson EM, Rossenu S, Karlsson MO.. Exposure-safety analysis of QTc interval and transaminase levels following bedaquiline administration in patients with drug-resistant tuberculosis. CPT Pharmacometrics Syst Pharmacol 2021; 10:153108–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Achour B, Barber J, Rostami-Hodjegan A.. Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos 2014; 42:1349–56. [DOI] [PubMed] [Google Scholar]
- 14. McLeay SC, P V, van Heeswijk RP, Green B.. Population pharmacokinetics of bedaquiline (TMC207), a novel antituberculosis drug. Antimicrob Agents Chemother 2014; 58:5315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Svensson EM, Dosne AG, Karlsson MO.. Population pharmacokinetics of bedaquiline and metabolite M2 in patients with drug-resistant tuberculosis: the effect of time-varying weight and albumin. CPT Pharmacometrics Syst Pharmacol 2016; 5:682–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization. Updates to the Tuberculosis (TB) Section of the Model List of Essential Medicines to align with formulation availability for TB programmes and current treatment recommendations in guidelines. https://www.who.int/selection_medicines/committees/expert/22/applications/6.2.4_new_TB_formulations.pdf. Accessed 9 February 2022. [Google Scholar]
- 17. Nahid P, Mase SR, Migliori GB, et al. . Treatment of drug-resistant tuberculosis. an official ATS/CDC/ERS/IDSA clinical practice guideline. Am J Respir Crit Care Med 2019; 200:e93–e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cholo MC, Steel HC, Fourie PB, Germishuizen WA, Anderson R.. Clofazimine: current status and future prospects. J Antimicrob Chemother 2012; 67:290–8. [DOI] [PubMed] [Google Scholar]
- 19. Nix DE, Adam RD, Auclair B, Krueger TS, Godo PG, Peloquin CA.. Pharmacokinetics and relative bioavailability of clofazimine in relation to food, orange juice and antacid. Tuberculosis (Edinb) 2004; 84:365–73. [DOI] [PubMed] [Google Scholar]
- 20. O’Donnell MR, Padayatchi N, Metcalfe JZ.. Elucidating the role of clofazimine for the treatment of tuberculosis. Int J Tuberc Lung Dis 2016; 20:52–7. [DOI] [PubMed] [Google Scholar]
- 21. Cholo MC, Mothiba MT, Fourie B, Anderson R.. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J Antimicrob Chemother 2017; 72:338–53. [DOI] [PubMed] [Google Scholar]
- 22. Feng PC, Fenselau CC, Jacobson RR.. Metabolism of clofazimine in leprosy patients. Drug Metab Disposit 1981; 9:521–4. [PubMed] [Google Scholar]
- 23. Holdiness MR. Clinical pharmacokinetics of clofazimine: a review. Clin Pharmacokinet 1989; 16:74–85. [DOI] [PubMed] [Google Scholar]
- 24. Brust JCM, Gandhi NR, Wasserman S, et al. . Effectiveness and cardiac safety of bedaquiline-based therapy for drug-resistant tuberculosis: a prospective cohort study. Clin Infect Dis 2021; 73:2083–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Svensson EM, Aweeka F, Park JG, Marzan F, Dooley KE, Karlsson MO.. Model-based estimates of the effects of efavirenz on bedaquiline pharmacokinetics and suggested dose adjustments for patients coinfected with HIV and tuberculosis. Antimicrob Agents Chemother 2013; 57:2780–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdelwahab MT, Wasserman S, Brust JCM, et al. . Clofazimine pharmacokinetics in patients with TB: dosing implications. J Antimicrob Chemother 2020; 75:3269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pandie M, Wiesner L, McIlleron H, et al. . Drug-drug interactions between bedaquiline and the antiretrovirals lopinavir/ritonavir and nevirapine in HIV-infected patients with drug-resistant TB. J Antimicrob Chemother 2016; 71:1037–40. [DOI] [PubMed] [Google Scholar]
- 28. Brill MJ, Svensson EM, Pandie M, Maartens G, Karlsson MO.. Confirming model-predicted pharmacokinetic interactions between bedaquiline and lopinavir/ritonavir or nevirapine in patients with HIV and drug-resistant tuberculosis. Int J Antimicrob Agents 2017; 49:212–7. [DOI] [PubMed] [Google Scholar]
- 29. Diacon AH, Dawson R, von Groote-Bidlingmaier F, et al. . Bactericidal activity of pyrazinamide and clofazimine alone and in combinations with pretomanid and bedaquiline. Am J Respir Crit Care Med 2015; 191:943–53. [DOI] [PubMed] [Google Scholar]
- 30. Ahmad S, Ghosh A, Nair DL, Seshadri M.. Simultaneous extraction of nuclear and mitochondrial DNA from human blood. Genes Genet Syst 2007; 82:429–32. [DOI] [PubMed] [Google Scholar]
- 31. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taliun D, Harris DN, Kessler MD, et al. . Sequencing of 53 831 diverse genomes from the NHLBI TOPMed Program. Nature 2021; 590:290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. UCSC Genome Browser. liftOver. http://genome.ucsc.edu/cgi-bin/hgLiftOver. Accessed 9 February 2022.
- 34. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D.. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet 2006; 38:904–9. [DOI] [PubMed] [Google Scholar]
- 35. Clarke L, Fairley S, Zheng-Bradley X, et al. . The International Genome Sample Resource (IGSR): a worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res 2017; 45:D854–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. PharmGKB. The Pharmacogenomics Knowledgebase. https://www.pharmgkb.org/. Accessed 3 February 2021.
- 37. NHGRI-EBI. GWAS Catalog—the NHGRI-EBI Catalog of published genome-wide association studies. https://www.ebi.ac.uk/gwas/. Accessed 3 February 2021.
- 38. Pruim RJ, Welch RP, Sanna S, et al. . LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2010; 26:2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hustert E, Haberl M, Burk O, et al. . The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics 2001; 11:773–9. [DOI] [PubMed] [Google Scholar]
- 40. Lamba J, Hebert JM, Schuetz EG, Klein TE, Altman RB.. PharmGKB summary: very important pharmacogene information for CYP3A5. Pharmacogenet Genomics 2012; 22:555–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khan AR, Raza A, Firasat S, Abid A.. CYP3A5 gene polymorphisms and their impact on dosage and trough concentration of tacrolimus among kidney transplant patients: a systematic review and meta-analysis. Pharmacogenomics J 2020; 20:553–62. [DOI] [PubMed] [Google Scholar]
- 42. National Center for Biotechnology Information. dbSNP—short genetic variations. https://www.ncbi.nlm.nih.gov/snp/rs776746. Accessed 23 December 2021.
- 43. Yoshihara Y, Kawasaki M, Tamada A, Nagata S, Kagamiyama H, Mori K.. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol 1995; 28:51–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.