Abstract
Background
Infiltration is important for the surgical planning and prognosis of pituitary adenomas. Differences in preoperative diagnosis have been noted. The aim of this article is to assess the accuracy of machine learning analysis of texture-derived parameters of pituitary adenoma obtained from preoperative MRI for the prediction of high infiltration.
Methods
A total of 196 pituitary adenoma patients (training set: n = 176; validation set: n = 20) were enrolled in this retrospective study. In total, 4120 quantitative imaging features were extracted from CE-T1 MR images. To select the most informative features, the least absolute shrinkage and selection operator (LASSO) and variance threshold method were performed. The linear support vector machine (SVM) was used to fit the predictive model based on infiltration features. Furthermore, the receiver operating characteristic curve (ROC) was generated, and the diagnostic performance of the model was evaluated by calculating the area under the curve (AUC), accuracy, precision, recall, and F1 value.
Results
A variance threshold of 0.85 was used to exclude 16 features with small differences using the LASSO algorithm, and 19 optimal features were finally selected. The SVM models for predicting high infiltration yielded an AUC of 0.86 (sensitivity: 0.81, specificity 0.79) in the training set and 0.73 (sensitivity: 0.87, specificity: 0.80) in the validation set. The four evaluation indicators of the predictive model achieved good diagnostic capabilities in the training set (accuracy: 0.80, precision: 0.82, recall: 0.81, F1 score: 0.81) and independent verification set (accuracy: 0.85, precision: 0.93, recall: 0.87, F1 score: 0.90).
Conclusions
The radiomics model developed in this study demonstrates efficacy for the prediction of pituitary adenoma infiltration. This model could potentially aid neurosurgeons in the preoperative prediction of infiltration in PAs and contribute to the selection of ideal surgical strategies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s41016-022-00290-4.
Keywords: Pituitary adenoma, Machine learning, Preoperative prediction, Magnetic resonance imaging, Infiltration
Background
Pituitary adenomas are common central nervous system tumors, accounting for approximately 15–20% of all intracranial tumors, with an incidence of 80–90/100,000 [1, 2]. Invasive pituitary adenomas account for approximately 15% of all types of pituitary adenomas. Because these tumors often invade the cavernous sinus, sphenoid sinus, internal carotid artery and suprasellar space, it is more difficult to surgically remove invasive pituitary adenomas compared with noninvasive pituitary adenomas. Moreover, the total resection rate is low, and these tumors easily relapse after surgery and have a poor prognosis [3, 4]. When pituitary adenomas are classified as Knosp grade 3 or 4 on coronal MRI, it has become a common practice to mark them as highly aggressive, and reports have clearly noted that a high Knosp grade (> grade 3) and high recurrence risk are related [5, 6]. In addition, an increasing number of studies use a high Ki67 proliferation index (>3%) as evidence of invasion and proliferation of invasive pituitary adenomas and to distinguish invasive pituitary adenomas from noninvasive pituitary adenomas [7, 8]. However, the Ki67 proliferation index is obtained by immunohistochemical analysis of postoperative pathology.
MRI is the best method for imaging diagnosis of pituitary adenomas. Given the fact that functional pituitary adenomas and nonfunctional pituitary adenomas are initially captured by MRI, the treatment plan is different [9–11]. Therefore, when the brain MRI examination reveals a pituitary adenoma, especially when the tumor is small, if noninvasive MRI can be exclusively used to identify the function of the pituitary adenoma before the operation, this methodology can further identify the high invasiveness and height of the pituitary adenoma before the operation. However, the human eye diagnosis of diagnostic imaging physicians is limited, and it is impossible to identify more subtle tumor features with the naked eye. The emergence of radiomics can precisely help solve this problem.
Radiomics is an emerging field designed to extract a large number of high-dimensional scalable features from medical imaging data (possibly combined with clinical or genomic data) and establish relevant statistical models to assist in diagnostic, prognostic, and therapeutic monitoring. The radiomics workflow includes imaging, ROI segmentation, feature extraction and analysis. Then, statistical models are subsequently designed based on machine learning algorithms that must be adjusted according to clinical or biological issues and the prior knowledge available [12].
This report proposes a method of using radiomics to classify MRI images of pituitary tumors based on infiltration. We aim to use machine learning algorithms to develop a radiomics model that predicts the high invasiveness of pituitary adenomas, providing a new preoperative evaluation method for patients with pituitary adenomas to better formulate treatment strategies.
Methods
Patients
Ethical approval for this retrospective analysis was obtained from the science research ethics committee of Linyi People’s Hospital, and the need for informed consent was waived. All patients with pituitary tumors who underwent surgical resection at our hospital from February 2016 to July 2020 were enrolled.
We collected clinical information of patients, including age, sex, preoperative computer MRI images, bilateral Knosp classification, preoperative hormones, endocrine symptoms, postoperative pathology, and immunohistochemical results. According to the relationship between cavernous sinus and the adenoma during operation, we define invasiveness tumors as highly aggressive and others as not highly aggressive. A Ki67 proliferation index > 3% was defined as a high Ki67 proliferation index [5, 7]. The following inclusion criteria were employed: (1) the quality of preoperative computer MRI images is good, images lack artifacts, and the images are from the same MRI scanner; (2) complete preoperative hormone examination; and (3) all patients were pathologically confirmed as having pituitary adenomas based on immunohistochemistry. The following exclusion criteria were applied: (1) patients who received previous surgery, drugs, or radiotherapy, and (2) preoperative computer MRI images have obvious artifacts. A dataset containing 196 patients was obtained (93 males and 103 females, average age: 52 ± 12.98 years, age range 11–76 years). The dataset is divided into three scan bits, including coronal plane, transverse plane, and sagittal plane, with 198 samples of coronal plane and sagittal plane scans and 196 transverse plane samples. The training set (n=176) and the validation set (n=20) are divided in a ratio of 9:1. Table 1 lists the specific characteristics of the patients in the training set and validation set. The training set is used for image feature construction and model development, and the validation set is used for model verification. Our flow chart is shown in Fig. 1.
Table 1.
Patients clinical characteristics (n=196)
| Characteristic | Training set (n=176) | Validation set (n=20) | Whole set (n=196) | P-value |
|---|---|---|---|---|
| Age (year, mean ± std) | 52.43 ± 12.82 | 48.70 ± 14.05 | 51.99 ± 12.98 | 0.224 |
| Gender | 0.809 | |||
| Male | 83 (47.16%) | 10 (50.00%) | 93 (47.45%) | |
| Female | 93 (52.84%) | 10 (50.00%) | 103 (52.55%) | |
| Knosp grade | 0.608 | |||
| Grades 0–2 | 81 (46.02%) | 8 (40.00%) | 89 (45.41%) | |
| Grades 3–4 | 95 (53.98%) | 12 (60.00%) | 107 (54.59%) | |
| Hormone hypersecreting tumors | 0.707 | |||
| Yes | 87 (49.43%) | 9 (45.00%) | 96 (48.98%) | |
| No | 89 (50.57%) | 11 (55.00%) | 100 (51.02%) | |
| Infiltration | 0.419 | |||
| High invasion | 95 (53.98%) | 12 (60%) | 107 (60.79%) | |
| Not high invasion | 81 (46.02%) | 8 (40%) | 99 (39.21%) | |
| Ki67 proliferation index | 0.103 | |||
| < 3% | 120 (68.18%) | 10 (50.00%) | 130 (66.33%) | |
| ≥ 3% | 56 (31.82%) | 10 (50.005) | 66 (33.67%) |
std standard deviation. P value < 0.5 represents a significant difference
Fig. 1.
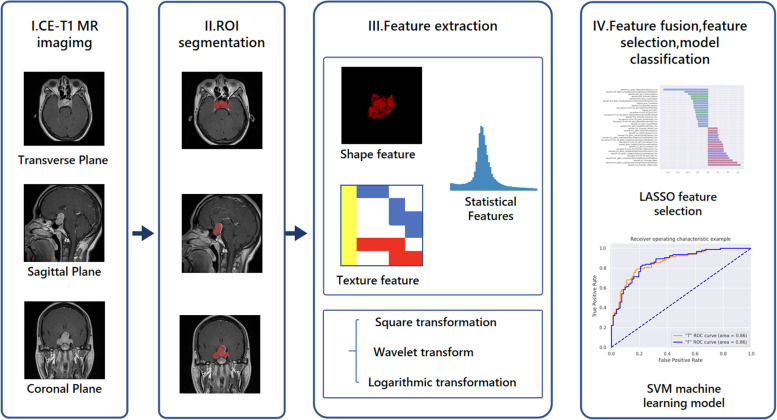
Flow chart of this study. I Three scanning positions of the original CE-T1 image: axial, sagittal, and coronal. II Segmentation of ROI. III Transform after extracting features from ROI. IV Feature selection and model establishment
MRI image acquisition
All patients underwent saddle area scans using the same MRI scanner (Siemens, 3.0T, Trio, Germany) in our hospital. The imaging schemes included T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), and CE-T1. Given that the image resolution of each slice that CE-T1 may obtain is very high, the contrast difference between normal tissue and tumor tissue will increase after the injection of the contrast agent [13]. Therefore, in this study, we selected the patient images from the archive system in our hospital. The CE-T1 image is used for analysis. The collection and setting parameters of the CE-T1 sequence are as follows: repetition time/echo time (TR/TE) = 232/8.1 ms, slice thickness=2 mm, field of view (FOV) = 200 × 200 mm2, and voxel size = 0.9 × 0.6 × 2.0 mm3.
Tumor segmentation
After the MRI image was examined by a radiologist, the ROI of all tumors was segmented by two neurosurgeons and radiologists with more than 10 years of experience in pituitary adenoma diagnosis and treatment. Two radiologists (experts 1 and 2 with 11 and 27 years of experience in brain MRI, respectively) delineated ROIs while blinded to the pathology results using 3D Slicer (https://www.slicer.org). Intersection over union (IOU) was calculated to determine the agreement between two radiologists, thus evaluating interobserver reproducibility. For cases with an IOU greater than 0.8, we chose the delineation result from expert 1 as a reference, and the ROIs of the remaining cases were then determined by a third experienced radiologist. Under double-blind conditions, 3D Slicer software was used to analyze the coronal position (COR), and sagittal (SAG) and axial (TRA) CE-T1 images were manually depicted. Disagreements arising during the analysis were resolved by the two physicians through negotiation. Finally, 588 ROIs were divided from 196 coronal, sagittal, and axial samples for subsequent radiomic analysis. In addition, the two physicians evaluated the Knosp classification of all samples in the coronal image under double-blind conditions, and the results were consistent.
Feature dimension reduction
Image value processing: The voxels of the MRI image exhibit large variations over the numerical range. Therefore, it is particularly important to reduce the variation of the dataset MRI images. The image value processing method used in this classification task is as follows: Assuming that the set of all voxel values of an image is X, the 99% quantile P of pixel values in the image region is taken, and the image scaling value M is defined. The pixel values in the image are calculated based on the following equation: window area = (X·M)/P. In addition, the Xwin voxel value greater than M is equal to M.
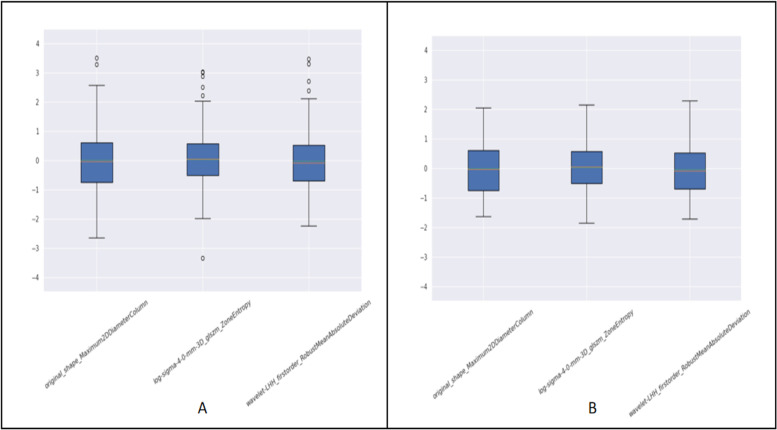
Processing outlier points: When counting a feature of all samples, some feature points deviate far from the overall distribution, which affects the classification effect of subsequent models, so these outlier points are treated as follows. Assuming that a set of a feature of the whole sample is F, to take the 99.9% quantile P 99.9, the F is set to a value greater than P 99.9 values, and values are all equal to P 99.9. The eigenvalue point distribution of the nonprocessed data (Fig. 2A) versus the processed data (Fig. 2B) is described as follows:
Resampling: To unify the spacing of all images to [0.625, 0.625, 2.4].
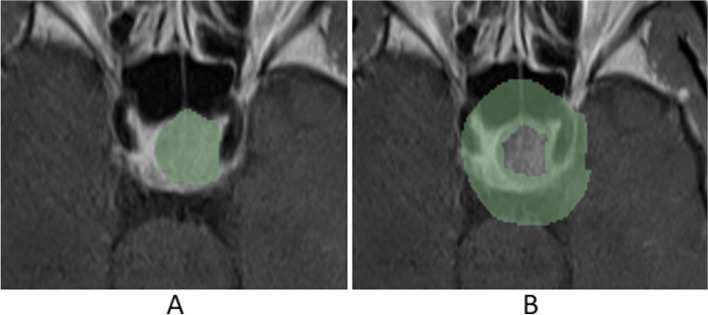
Multiple ROIs: For this classification task, certain pathologies associated with infiltration were present in the surrounding areas of the tumor; therefore, the task adopted a mask in the original tumor region. The mask from the area surrounding the tumor was also used to obtain this mask. Using the SimpleITK kit or Opencv kit, corrosion operations were performed on the original mask. Mask_e was obtained to expand the original mask, and mask_d was obtained. By subtracting mask_e from mask_d, the mask of the area surrounding the tumor was obtained (Fig. 3).
Fig. 2.
The contrast of handles outlier points. A The distribution of does not process outlier point. B The distribution of processes outlier point
Fig. 3.
Mask of the area surrounding the tumor. A Mask ROI. B mask_edge ROI
Preprocessing used for classification task
We used a preprocessing method for the infiltration classification task (Additional file 1).
Feature extraction
Quantitative image features were extracted from MRI images using the Pyradiomics library (wavelet parameters: rbio1.1, LoG parameters for image value processing: original-mask sigma = 1.0, 2.0, 3.0, 4.0, 5.0, and 6.0; edge-mask sigma = 1.0, 2.0, 3.0, 4.0, 5.0, and 6.0; LoG parameters for no image value processing: original-mask sigma=2.0, 3.0, 4.0, 5.0; edge-mask sigma=1.0, 2.0, 3.0, 4.0, 5.0, and 6.0) in Additional file 2.
Feature dimension reduction
As described above, a large number of image features can be obtained. However, most of these features may be of no use for classification tasks. Therefore, it is necessary to reduce the features and identify features with high pathological correlation. To reduce redundancy, feature reduction was performed using the following methods:
The variance threshold method removes the eigenvalues with a variance less than the threshold. Using the LASSO model, L1 is regularized as a cost function with alphas=0.001 and a maximum of 10000 iterations.
Statistical analysis (classifier)
After feature screening, there are multiple supervised learning classifiers available for classification analysis based on the selected features. In this study, a model based on Radiomics is constructed using a linear support vector machine (LinearSVM) classifier, and the effectiveness of the model is verified. Parameter settings for the linear support vector machine LinearSVM are noted as follows: infiltration: C = 1, random_state = 23, and max_iter = 10000. To evaluate predictive performance, the operating feature (ROC) curve and curve area (AUC) were used in the training and validation datasets. The study uses five indicators to evaluate the performance of the classifiers: (1) accuracy (ACC), (2) precision (P) ((precision = true positives/(true positives + false-positives)), (3) Recall ® (recall = true positives/(true positives + false negatives)), (4) F1 Score (f1 score = P*R*2/(P + R)), and (5) support (the total number of samples in the test set).
Results
Clinical characteristics
A total of 196 patients (age, 51.99 ± 12.98 years) were enrolled in this study. The characteristics of the patients and tumors are shown in Table 1. No significant differences for all clinic-radiological factors (p = 0.103-0.809) were found between the training set and validation set, thereby justifying the use of the training set and validation set.
Statistical results of the datasets
The label distribution of the validation sets and training sets for different classification tasks is listed in Table 1. In total, 10% of all cases (male to female ratio: 10:10) were included in the validation set, and 90% of all cases (male to female ratio: 83:93) were included in the training set. In the validation set, 12 cases were highly invasive, accounting for 60% of all cases. Eight cases were not highly invasive, accounting for 40% of all cases. In the training test, 95 cases were highly invasive, accounting for 54% of all cases, and 81 cases were not highly invasive, accounting for 46% of all cases.
Feature screening results
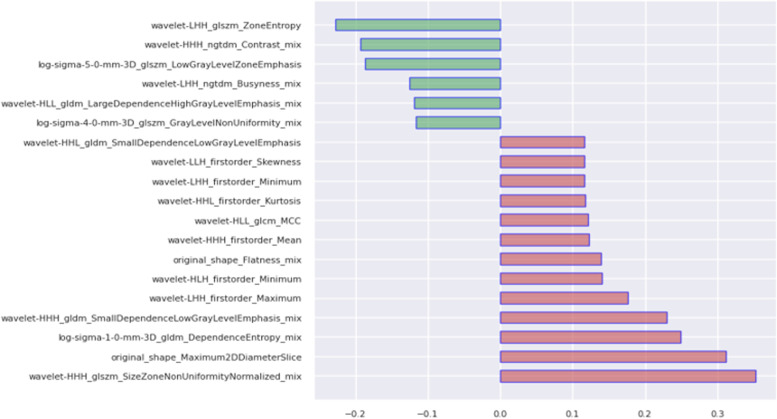
A total of 4120 quantitative imaging features were extracted from the three scanning positions of contrast-enhanced T1-weighted imaging (CE-T1). For highly invasive classification tasks, a variance threshold of 0.85 was used to exclude 16 features with small differences using the LASSO algorithm (Fig. 4), and 19 optimal features were finally selected (Table 2). The following lasso parameter settings were employed: alphas=0.001, max_iter (10000).
Fig. 4.
Weights obtained by lasso method in the classification task with high infiltration
Table 2.
Features selected for validation classification task
| Radiomic feature | Radiomic class | Filter | Mask |
|---|---|---|---|
| Maximum2DDiameterSlice | shape | original | original |
| LowGrayLevelZoneEmphasis | glszm | log-sigma-5-0 | original |
| Skewness | firstorder | wavelet-LLH | original |
| Maximum | firstorder | wavelet-LHH | original |
| Minimum | firstorder | wavelet-LHH | original |
| ZoneEntropy | glszm | wavelet-LHH | original |
| MCC | glcm | wavelet-HLL | original |
| Minimum | firstorder | wavelet-HLH | original |
| Kurtosis | firstorder | wavelet-HHL | original |
| SmallDependenceLowGrayLevelEmphasis | gldm | wavelet-HHL | original |
| Mean | firstorder | wavelet-HHH | original |
| Flatness | shape | original | egde |
| DependenceEntropy | gldm | log-sigma-1-0 | egde |
| GrayLevelNonUniformity | glszm | log-sigma-4-0 | egde |
| Busyness | ngtdm | wavelet-LHH | egde |
| LargeDependenceHighGrayLevelEmphasis | gldm | wavelet-HLL | egde |
| SizeZoneNonUniformityNormalized | glszm | wavelet-HHH | egde |
| Contrast | ngtdm | wavelet-HHH | egde |
| SmallDependenceLowGrayLevelEmphasis | gldm | wavelet-HHH | egde |
Features selected for validation classification task
Model performance
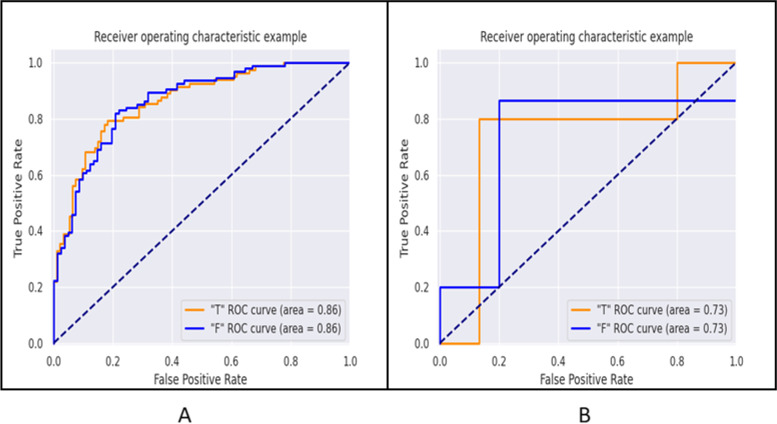
Figure 5 shows the ROC curves of the classification task. Table 3 shows the AUC, sensitivity, and specificity of different classification tasks in the training set and validation set (“T” represents “Yes,” “F” represents “None”). The radiomics model showed good discrimination. For highly invasive classification tasks, the AUC of the training set was 0.86 (95% confidence interval: 0.75–1.00), the sensitivity was 0.81, and the specificity was 0.79. The AUC of the validation set was 0.73 (95% confidence interval: 0.53–0.94), the sensitivity was 0.87, and the specificity was 0.80. The results show that the radiomics model we established is feasible for distinguishing highly aggressive pituitary adenomas.
Fig. 5.
ROC curve of high infiltration classification task. A Training set. B Validation set (“T” stands for “Yes,” “F” stands for “None”)
Table 3.
Results of ROC curve analysis and five indicators in the training set and verification set
| Training set | Verification set | |||
|---|---|---|---|---|
| Indicators | Infiltration | Indicators | Infiltration | |
| T | AUC | 0.86 | AUC | 0.73 |
| 95% CI | 0.75–1.00 | 95% CI | 0.53–0.94 | |
| Sensitivity | 0.81 | Sensitivity | 0.87 | |
| Specificity | 0.79 | Specificity | 0.80 | |
| Accuracy | 0.8 | Accuracy | 0.85 | |
| Precision | 0.82 | Precision | 0.93 | |
| Recall | 0.81 | Recall | 0.87 | |
| F1-score | 0.81 | F1-score | 0.90 | |
| Support | 94 | Support | 15 | |
| F | AUC | 0.86 | AUC | 0.73 |
| 95% CI | 0.75–1.00 | 95% CI | 0.53–0.94 | |
| Sensitivity | 0.79 | Sensitivity | 0.80 | |
| Specificity | 0.81 | Specificity | 0.87 | |
| Accuracy | 0.8 | Accuracy | 0.85 | |
| Precision | 0.78 | Precision | 0.67 | |
| Recall | 0.79 | Recall | 0.80 | |
| F1-score | 0.79 | F1-score | 0.73 | |
| Support | 82 | Support | 5 | |
The classifier evaluation results
We summarized the four evaluation indicators of the prediction model (accuracy rate, precision rate, recall rate, and F1 value). As shown in Table 3, the model achieved good diagnostic capabilities in the training set and independent verification set. Specifically, the accuracy was 0.80, the precision was 0.82, the recall was 0.81, and the F1 score was 0.81 in the training set. In addition, the accuracy was 0.85, the precision was 0.93, the recall was 0.87, and the F1 score was 0.90 in the independent verification set.
Discussion
Based on MRI, this study used radiomics to develop a radiomics model that predicts the high invasiveness of pituitary adenomas and achieved good results. The results demonstrate that the radiomics model predicts the invasiveness of pituitary adenomas. The model exhibits good potential for predicting highly invasive pituitary adenomas. This information may help clinicians assess the nature of the tumor before surgery and better formulate treatment strategies.
The concept of radiomics was first proposed by Dutch scholar Philippe Lambin and has been applied to the prediction and evaluation of a variety of tumors, such as colorectal cancer, lung cancer, and prostate cancer [14–19]. In recent years, radiomics has also been confirmed to exhibit increasing potential in the preoperative diagnosis of pituitary adenomas. The radiomics model established by Zhang et al. is used to distinguish subtypes of nonfunctional pituitary adenomas before surgery, and the model performs well [2]. Fan et al. applied radiomics to predict the response of patients with acromegaly to radiotherapy [20]. Niu et al. developed a radiomics model that predicted the invasion of pituitary adenomas on the cavernous sinus [21]. Peng et al. [22] developed a machine learning model for predicting the immunohistochemical classification of pituitary adenomas before surgery.
In this study, based on preoperative CE-T1 images of patients with pituitary adenomas and using machine learning methods, we developed a radiomics model that predicts the high invasiveness of pituitary adenomas. The good AUC value of the final training set and the validation set demonstrated its potential value in the prediction of high invasiveness of pituitary adenoma, and its reliability was verified by calculating the index of the classifier. Niu et al. identified a radiomics nomogram based on CE-T1 and T2 MR images for the individualized evaluation of cavernous sinus invasion in 194 patients with PAs (Knosp grades two or three) [21]. The radiomics model yielded area under the curve (AUC) values of 0.852 and 0.826 for the training and test sets, respectively. In contrast, our study enrolled patients to predict high invasiveness (cavernous sinus invasion). We calculated the area under the curve (AUC), accuracy, precision, recall, and F1 value to evaluate the diagnostic performance, but the performance of the two models needs to be further compared.
A very important issue in the application of radiomics is the selection of the best features. Unlike some previous studies [15, 16, 18, 23, 24], our research uses some innovative methods. First, in the application of image data, our study takes into account the clinical domain knowledge. For ROI segmentation, three window positions, including coronal, axial, and sagittal, were included to minimize errors and improve three-dimensional features. Second, regarding the microenvironment around the tumor, a mask expansion method is used when preprocessing the data. Third, in the feature extraction method, filter transformation is used in addition to the original features to extract finer texture features. These results laid the foundation for the good performance of the model.
Our research has some limitations. First, our research was conducted at a single center. All image data were obtained from the same machine in a hospital. Whether our model is suitable for multicenter research remains to be demonstrated. In the future, we need to test the imaging data from different centers to validate our model. In addition, all ROI annotations are performed manually, which is time-consuming. Given the availability of more sample sets in the future, if an automatic segmentation algorithm can be created, an end-to-end application can be formed. Therefore, we need to identify an accurate automatic segmentation algorithm. Third, research has revealed that the consistency of T2-weighted images may be better25. In our study, only CE-T1 images were used, and no comparison with other sequences was performed.
Conclusion
In summary, we used radiomics to create a predictive model of highly invasive pituitary adenomas, and independently verified its reliability in the validation set. In the future, the number of samples will be increased, and the model will be continuously optimized. The diagnostic ability of the model is expected to be further improved, and this model may help neurosurgeons formulate better treatment strategies.
Supplementary Information
Acknowledgements
The authors would like to thank the Ebond (Beijing) Intelligence Technology Co., Ltd., for proofreading of the manuscript.
Abbreviations
- ACC
Accuracy
- AIC
Akaike’s information criterion
- AUC
Area under the curve
- BIC
Bayesian information criterion
- CE-T1
Contrast-enhanced T1-weighted
- CI
Confidence interval
- CS
Cavernous sinus
- DCA
Decision curve analysis
- FOV
Field of view
- LASSO
Least absolute shrinkage and selection operator
- MRI
Magnetic resonance imaging
- PAs
Pituitary adenomas
- ROC
Receiver operating characteristic curve
- ROIs
Regions of interests
- SSA
Somatostatin analogs
- SVM
Support vector machine
- T1WI
T1-weighted imaging
- TR/TE
Repetition time/echo time
Authors’ contributions
MW and CZ conceived and designed the project. Each author has contributed significantly to the submitted work. CZ drafted the manuscript. All authors contributed to the preparation and revision of the manuscript. All authors read and approved the final manuscript.
Funding
The present study was supported by funds of Postdoctoral Innovation Program of Shandong Province (NO. 202103064). Linyi People’s Hospital Doctoral Research Foundation (NO. 2021LYBS05).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Approval of Ethics Committee comments of the Linyi People’s Hospital was obtained before commencing the study.
Consent for publication
All authors consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lim CT, Korbonits M. Update on the clinicopathology of pituitary adenomas[J] Endocr Pract. 2018;24(5):473–488. doi: 10.4158/EP-2018-0034. [DOI] [PubMed] [Google Scholar]
- 2.Zhang S, Song G, Zang Y, et al. Non-invasive radiomics approach potentially predicts non-functioning pituitary adenomas subtypes before surgery[J] Eur Radiol. 2018;28(9):3692–3701. doi: 10.1007/s00330-017-5180-6. [DOI] [PubMed] [Google Scholar]
- 3.De Sousa SMC, McCormack AI, et al. Aggressive pituitary tumors and pituitary carcinomas. In: Feingold KR, Anawalt B, Boyce A, et al., editors. Endotext. South Dartmouth (MA): MDText.com, Inc; 2018. [Google Scholar]
- 4.Lopes MBS. The 2017 World Health Organization classification of tumors of the pituitary gland: a summary[J] Acta Neuropathol. 2017;134(4):521–535. doi: 10.1007/s00401-017-1769-8. [DOI] [PubMed] [Google Scholar]
- 5.Ilie MD, Jouanneau E, Raverot G. Aggressive pituitary adenomas and carcinomas[J] Endocrinol Metab Clin North Am. 2020;49(3):505–515. doi: 10.1016/j.ecl.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Yuhan L, Zhiqun W, Jihui T, Renlong P. Ki-67 labeling index and Knosp classification of pituitary adenomas. Br J Neurosurg. 2021:1–5. [DOI] [PubMed]
- 7.Lamback EB, Guterres A, Barbosa MA, et al. Cyclin A in nonfunctioning pituitary adenomas[J] Endocrine. 2020;70(2):380–387. doi: 10.1007/s12020-020-02402-5. [DOI] [PubMed] [Google Scholar]
- 8.Moldovan IM, Şuşman S, Pîrlog R, et al. Molecular markers in the diagnosis of invasive pituitary adenomas - an immunohistochemistry study[J] Rom J Morphol Embryol. 2017;58(4):1357–1364. [PubMed] [Google Scholar]
- 9.Møller MW, Andersen MS, Glintborg D, Pedersen CB, Halle B, Kristensen BW, Poulsen FR. [Pituitary adenoma]. Ugeskr Laeger. 2019;181(20):V05180331. [PubMed]
- 10.Ježková J, Marek J, Vaněčková M. Treatment of pituitary adenomas. Casopis Lekaru Ceskych. 2017;156(5):266–270. [PubMed] [Google Scholar]
- 11.Lake MG, Krook LS, Cruz SV. Pituitary adenomas: an overview[J] Am Fam Physician. 2013;88(5):319–327. [PubMed] [Google Scholar]
- 12.Liu Z, Wang S, Dong D, et al. The applications of radiomics in precision diagnosis and treatment of oncology: opportunities and challenges[J] Theranostics. 2019;9(5):1303–1322. doi: 10.7150/thno.30309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonneville JF. Magnetic resonance imaging of pituitary tumors. Front Horm Res. 2016;45:97–120. doi: 10.1159/000442327. [DOI] [PubMed] [Google Scholar]
- 14.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non- small cell lung cancer. Radiology. 2016;281(3):947–957. doi: 10.1148/radiol.2016152234. [DOI] [PubMed] [Google Scholar]
- 17.Shi L, Rong Y, Daly M, et al. Cone-beam computed tomography-based delta-radiomics for early response assessment in radiotherapy for locally advanced lung cancer. Phys Med Biol. 2020;65(1):015009. doi: 10.1088/1361-6560/ab3247. [DOI] [PubMed] [Google Scholar]
- 18.Shiradkar R, Podder TK, Algohary A, Viswanath S, Ellis RJ, Madabhushi A. Radiomics based targeted radiotherapy planning (Rad-TRaP): a computational framework for prostate cancer treatment planning with MRI. Radiat Oncol. 2016;11(1):148. doi: 10.1186/s13014-016-0718-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Ford JC, Dogan N, et al. Magnetic resonance imaging (MRI)-based radiomics for prostate cancer radiotherapy. Transl Androl Urol. 2018;7(3):445–458. doi: 10.21037/tau.2018.06.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y, Chai Y, Li K, et al. Non-invasive and real-time proliferative activity estimation based on a quantitative radiomics approach for patients with acromegaly: a multicenter study. J Endocrinol Invest. 2020;43(6):755–765. doi: 10.1007/s40618-019-01159-7. [DOI] [PubMed] [Google Scholar]
- 21.Niu J, Zhang S, Ma S, et al. Preoperative prediction of cavernous sinus invasion by pituitary adenomas using a radiomics method based on magnetic resonance images. Eur Radiol. 2019;29(3):1625–1634. doi: 10.1007/s00330-018-5725-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng A, Dai H, Duan H, et al. A machine learning model to precisely immunohistochemically classify pituitary adenoma subtypes with radiomics based on preoperative magnetic resonance imaging. Eur J Radiol. 2020;125:108892. doi: 10.1016/j.ejrad.2020.108892. [DOI] [PubMed] [Google Scholar]
- 23.Wu J, Aguilera T, Shultz D, et al. Early-stage non-small cell lung cancer: quantitative imaging characteristics of (18) FFluorodeoxyglucose PET/CT allow prediction of distant metastasis. Radiology. 2016;281:270–278. doi: 10.1148/radiol.2016151829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie K, Shi LM, Chen Q, et al. Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res. 2016;22:5256–5264. doi: 10.1158/1078-0432.CCR-15-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.