Figure 1.
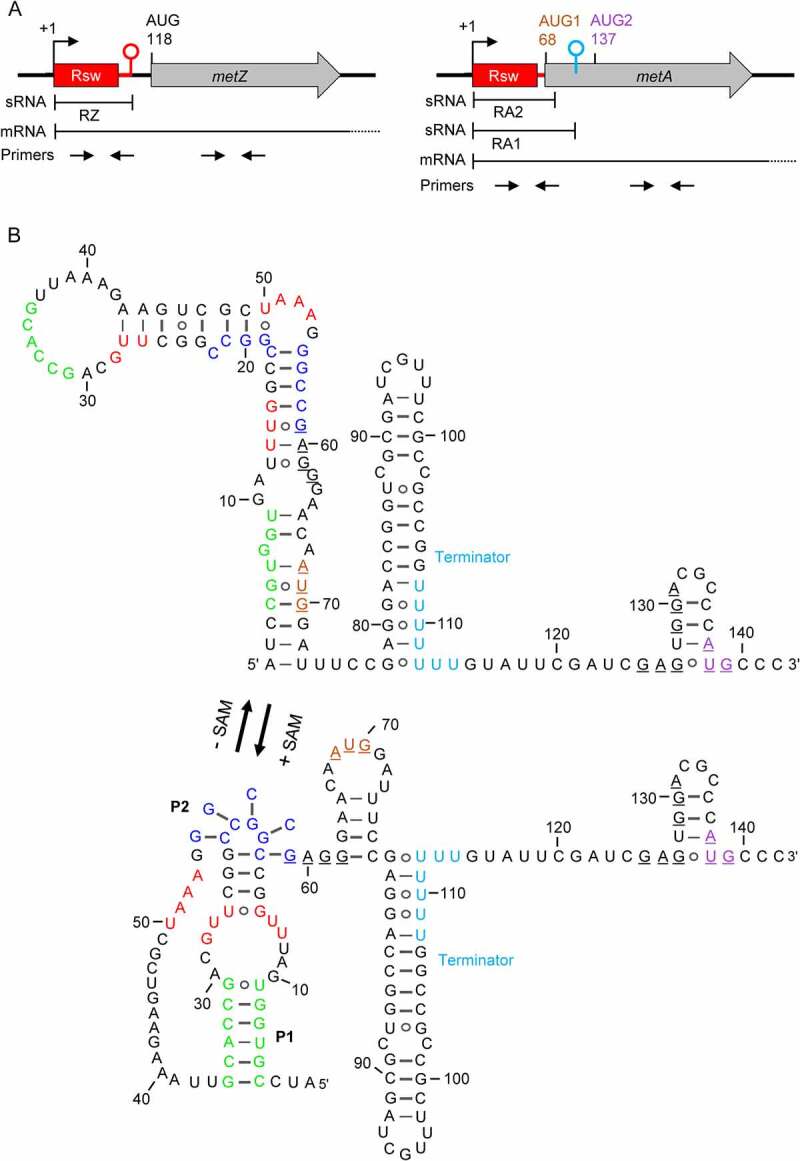
SAM-II riboswitches in S. meliloti 2011. A) Scheme of the metZ and metA genes and their relevant features. The metZ and metA ORFs are depicted by grey arrows. They are preceded by SAM-II riboswitches depicted in red. Rsw indicates the SAM-binding aptamer. Flexed arrows and hairpins show transcription start sites (TSSs at positions +1) and transcriptional terminators, respectively. Positions of adenine in (putative) AUG start codons are given. Regions corresponding to the riboswitch-containing sRNAs RZ, RA1 and RA2, and to the metZ and metA mRNAs are indicated (mRNA terminators are not shown). Indicated are also the positions of the primer pairs used for qRT-PCR analysis. B) The metA riboswitch and downstream sequences. Shown are the predicted alternative RNA structures adopted without SAM binding (-SAM) and upon interaction with SAM (+ SAM). Red nucleotides: the SAM-binding pocket (SBP); green nucleotides: sequences building stem 1 (P1) upon SAM binding; dark blue nucleotides: sequences building a pseudoknot (P2) upon SAM binding (according to [8]). The SD-like sequence and putative AUG start codons are underlined. AUG1 and AUG2 are coloured according to panel A). The U stretches in the transcriptional terminator between AUG1 and AUG2 are shown in cyan. The metZ riboswitch is shown in Fig. S1.
