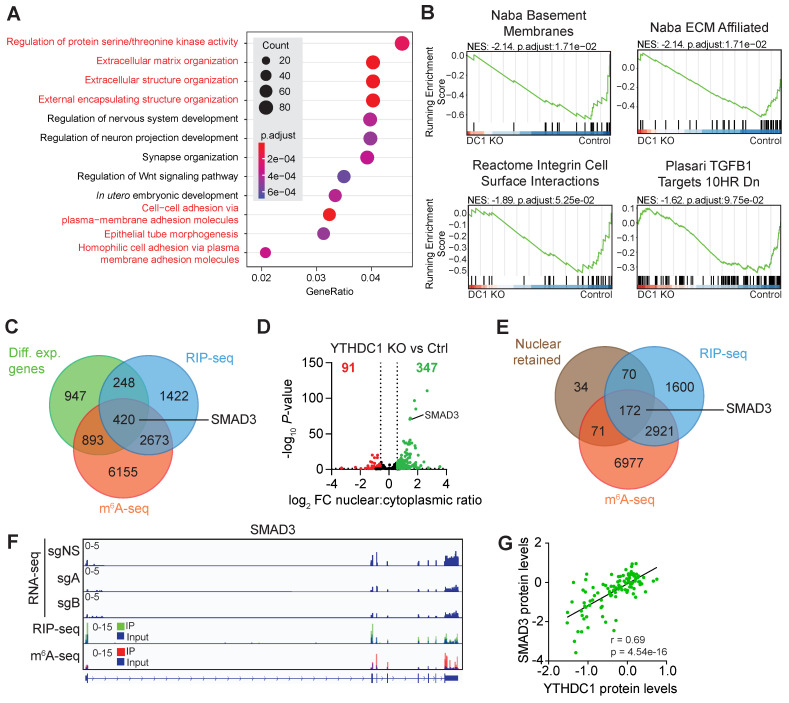
Figure 2.
Identification of YTHDC1 target mRNAs in MDA-MB-231 cells by integrated analysis of sequencing data. A) Gene Ontology (GO) analysis of significantly differentially expressed genes in MDA-MB-231 YTHDC1 KO cells compared to control. Pathways related to metastasis are highlighted in red. B) Pathways related to metastasis enriched in Gene Set Enrichment Analysis (GSEA) of significantly differentially expressed genes in MDA-MB-231 YTHDC1 KO cells compared to control. C) Venn diagram of differentially expressed genes overlapped with RIP-seq and m6A-seq data. D) Volcano plot showing changes in mRNA nuclear export following YTHDC1 KO compared to control. E) Venn diagram of nuclear retained transcripts overlapped with RIP-seq and m6A-seq data. F) RNA-seq, RIP-seq and m6A-seq tracks for SMAD3. G) Correlation between SMAD3 protein and YTHDC1 protein expression levels from CPTAC breast cancer tumor samples. Pearson's correlation test.