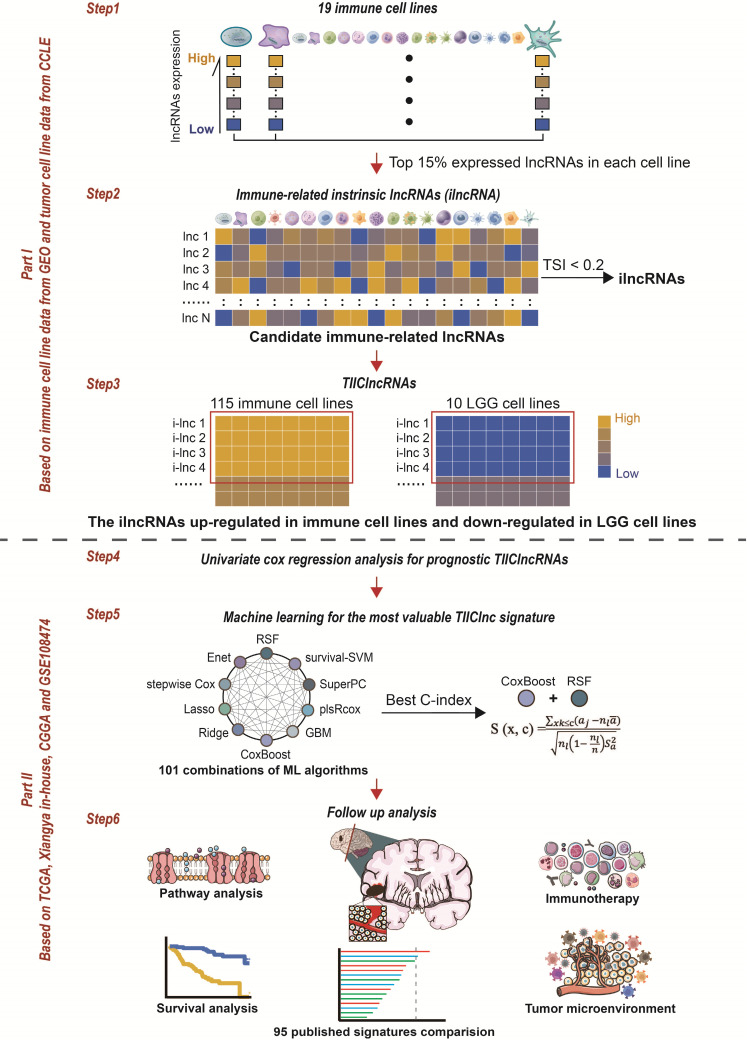
Figure 1.
The computational framework for establishing the TIIClnc signature. The top 15% expressed lncRNAs were taken for candidate immune-related lncRNAs for each immune cell line. TSI was applied to calculate the expression specificity of candidate immune-related lncRNAs for each cell type. The highly expressed lncRNAs in all immune cell types were identified as immune-related ilncRNA. ilncRNAs significantly upregulated in immune cell lines and downregulated in LGG cell lines were defined as TIIClncRNAs. Univariate cox regression analysis was further used to screen out the prognostic TIIClncRNAs. All combinations of 10 machine learning algorithms, including RSF, Enet, Lasso, Ridge, stepwise Cox, CoxBoost, plsRcox, SuperPC, GBM, and survival-SVM, based on a 10-fold cross-validation were further used to screen out the most valuable TIIClnc signature with the highest C-index. The TIIClnc signature was finally generated based on the combination of RSF and CoxBoost. The association between the TIIClnc signature, prognosis, tumor immune microenvironment, and immunotherapy response was comprehensively investigated.