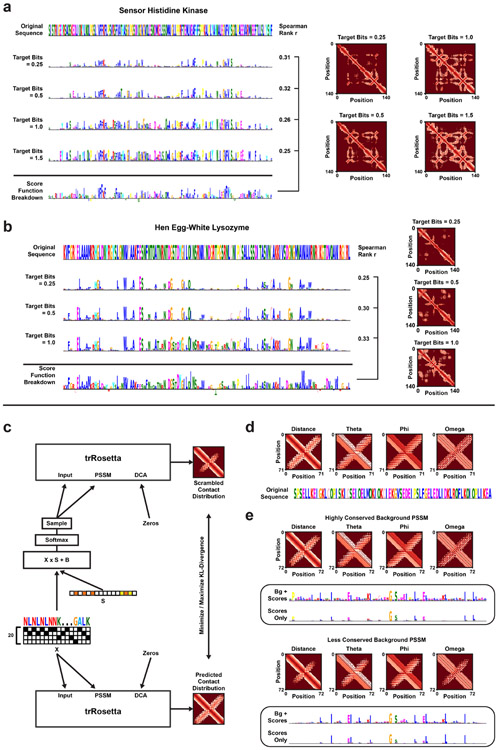
Extended Data Fig. 7.
(a) Four different Inclusion-PSSMs optimized to reconstruct the structural trRosetta prediction of a Sensor Histidine Kinase. Each PSSM is optimized for increasingly larger tbits. The bottom sequence logo represents the Rosetta score function breakdown per residue (−REU). Spearman r ranged between 0.25 and 0.32 when comparing the absolute numbers of Rosetta energy values to the optimized importance scores. Shown is also the average structure prediction for 512 samples. (b) Inclusion-Scrambled PSSMs of the Hen Egg-white Lysozyme. The PSSM was re-optimized for three different target conservation bits. Spearman r ranged between 0.25 and 0.33 compared to the Rosetta score function. (c) Architecture for per-example scrambling of a single protein sequence according to the contact distributions predicted by trRosetta. Here, we do not use a Multiple Sequence Alignment (MSA), but instead pass the Gumbel-sampled sequence to the PSSM input and an all-zeros matrix to the DCA input. Total KL-divergence between trRosetta-predicted distributions (distance and angle-grams) of the original sequence and samples drawn from the scrambled PSSM is either minimized or maximized (inclusion or occlusion respectively). (d) Reference sequence and predicted contact distribution for a hairpin protein engineered by Activation Maximization. (e) Top: Inclusion-PSSM of the engineered hairpin protein, obtained after optimization with a highly conserved background distribution based on the MSA. Bottom: Inclusion-PSSM of the engineered hairpin protein with a less conserved background distribution (smoothed with pseudo counts).