Abstract
SIRT1 and FOXO3 are both associated with longevity. Molecular biology research in many organisms (yeast, nematode worm Caenorhabditis elegans, and mice mammalian models) shows SIRT1 acts on the FOXO family of forkhead transcription factors to respond to oxidative stress better, shifting processes away from cell death toward stress resistance. Human population studies need epidemiologic evidence. We used an open cohort of 3 166 community-dwelling participants in China with follow-up from 2008 to 2018. The mean age at baseline was 84.6 years. In 16 375 person-years of follow-up, there were 1 968 mortality events. SIRT1 and FOXO3 exhibited Mendelian randomization as there was no correlation with each other and with baseline study population characteristics. Some SIRT1 and FOXO3 single-nucleotide polymorphisms showed protective effects for mortality risk. The FOXO3 protective effect was stronger in females, and the SIRT1 protective effect was stronger in male study participants. We did not see evidence of a synergistic effect of being carriers of both SIRT1 and FOXO3 advantageous alleles.
Keywords: Effect modification, FOXO3, Gene–gene interaction, Longevity, Sex difference, SIRT1
Forkhead box “O” 3 (FOXO3 or FOXO3A) and Sirtuin 1 (SIRT1) are genes associated with longevity. These genes may work together to regulate aging processes, with molecular biology evidence showing SIRT1 upregulates the FOXO family of Forkhead transcription factors to better respond to cellular stress, induce cell cycle arrest, resistance to oxidative stress, and aid in the insulin signaling pathway (1,2). These experimental findings were conducted in a variety of organisms including in yeast, the nematode worm Caenorhabditis elegans, and mice mammalian models (3). FOXO transcription factors and sirtuin deacetylases are critical regulators of mammalian vascular development and disease (4,5). Currently, no studies have assessed the combined effect of FOXO3 and SIRT1 on longevity and morbidity in population cohorts. We used a cohort of older Chinese to study the interaction of SIRT1 single-nucleotide polymorphisms (SNPs) and FOXO3 SNPs on mortality. First, we assess the individual effect of FOXO3 and SIRT1 on mortality. Second, we evaluate whether the effect of FOXO3 and SIRT1 varies by sex. Third, we assess the interaction term of FOXO3 and SIRT1 to look for synergistic effects.
Method
Study Population
We analyzed the Chinese Longitudinal Healthy Longevity Survey (CLHLS) study. This study collected information from the oldest-old population drawn from rural and urban regions in 23 out of 31 provinces in China. The first survey started in 1998, and there were new participants recruited to replace the deceased older adult during the follow-up surveys in 2000, 2002, 2005, 2008, 2011, 2014, and 2018. We included participants first interviewed in 2008/2009 and excluded those aged younger than 65, non-Han Chinese, without genetic data, and lost in the first follow-up. The final sample consisted of 3 166 participants.
Genotype Assessment of FOXO3 and SIRT1
The Beijing Genomics Institute performed the genotyping for 13 228 individuals using a customized chip for Chinese ancestry based on the previous CLHLS Genome-Wide Association Study (GWAS). The GWAS genotyping and quality control procedures were reported previously (6). The replication study targeted 27 656 longevity-phenotype-related SNPs. We extracted the same tagging FOXO3 and SIRT1 SNPs as previous longevity studies (7,8): FOXO3 rs4946936, FOXO3 rs2802292, FOXO3 rs2253310, SIRT1 rs12778366, SIRT1 rs3758391, SIRT1 rs2273773, and SIRT1 rs4746720.
We recoded the genotypes following the additive, heterozygote, minor-dominant, and minor-recessive models. The genotype that contains 0, 1, or 2 copies of minor allele was categorized as “AA,” “Aa,” and “aa.” In the additive model, we coded “AA” as 0 (reference group), “Aa” as 1, and “aa” as 2. In the heterozygote model, we coded “Aa” as 0 (reference group), “AA” as 1, and “aa” as 2. In the minor-dominant model, we coded “Aa”/“aa” as 1 and AA as 0. In the recessive model, we coded “aa” as 1 and “Aa”/“AA” as 0. In most analyses, carrying one copy of the minor allele “a” (additive model) is considered to have a decreased risk for mortality.
We further classified the combination of the genotype of FOXO3 and SIRT1 into 4 groups: carrying no minor allele of either FOXO3 or SIRT1; carrying at least one minor allele of FOXO3 and no minor allele of SIRT1; carrying no minor allele of FOXO3 and at least one minor allele of SIRT1; and carrying at least one minor allele of both FOXO3 and SIRT1.
Mortality Ascertainment
The next of kin reported the mortality information in the follow-up surveys between 2008 and 2018. The survival time was entered as month counted from the month of the initial interview to the month of death or censoring at the 2018 interview.
Statistical Analysis
We used a Cox proportional hazard model for every candidate SNP to evaluate their effect on mortality individually. We tested the interaction effect by adding the product of one FOXO3 SNP and one SIRT1 SNP. For each SNP, we draw the adjusted survival curve based on the expected survival curves calculated based on the Cox model separately for subpopulations (9). We further conducted stratified analyses by genotype and gender. We created new variables combining the FOXO3, SIRT1, and gender to compare the single and combined effects intuitively. All models were adjusted for age at baseline and gender. In the sensitivity analyses, we additionally adjusted for education, residence, marriage, exercise, smoking, drinking alcohol, and body mass index (BMI). We calculated hazard ratios (HRs) and 95% confidence intervals (CIs) to indicate associations between SNPs and mortality using R 4.0.0.
Results
We studied a total of 3 166 participants. During 16 375 person-years of follow-up, there were 1 968 mortality events. At baseline, the participants had a mean age of 85 (SD: 11), and 53% (n = 1 679) were female (Table 1). The distributions of FOXO3 and SIRT1 SNPs were not correlated with each other. We did not see meaningful or consistent trends in the difference of FOXO3 and SIRT1 SNP distributions by baseline age, years of education, urban or rural residence location, marital status, exercise frequency, smoking, alcohol consumption, and BMI measurement (Table 1; Supplementary Tables 1 and 2).
Table 1.
Population Characteristics at Baseline
| FOXO3 rs2802292 Minor Allele Number | SIRT1 rs3758391 Minor Allele Number | Overall (N = 3 166) | |||||
|---|---|---|---|---|---|---|---|
| 0 (N = 1 590) | 1 (N = 1 282) | 2 (N = 294) | 0 (N = 2 231) | 1 (N = 855) | 2 (N = 80) | ||
| Sex: n (%) | |||||||
| Male | 733 (46.1) | 602 (47.0) | 152 (51.7) | 1 041 (46.7) | 412 (48.2) | 34 (42.5) | 1 487 (47.0) |
| Female | 857 (53.9) | 680 (53.0) | 142 (48.3) | 1 190 (53.3) | 443 (51.8) | 46 (57.5) | 1 679 (53.0) |
| Age (year) | |||||||
| Mean (SD) | 84.6 (11.4) | 85.6 (11.2) | 85.0 (11.0) | 84.8 (11.4) | 85.8 (10.9) | 84.8 (10.9) | 85.0 (11.3) |
| Education year | |||||||
| Mean (SD) | 2.11 (3.40) | 2.07 (3.37) | 2.30 (3.45) | 2.13 (3.43) | 2.09 (3.33) | 1.86 (2.96) | 2.11 (3.39) |
| Missing | 4 (0.3) | 2 (0.2) | 1 (0.3) | 6 (0.3) | 1 (0.1) | 0 (0) | 7 (0.2) |
| Residence: n(%) | |||||||
| City/town | 525 (33.0) | 398 (31.0) | 98 (33.3) | 733 (32.9) | 257 (30.1) | 31 (38.8) | 1 021 (32.2) |
| Rural | 1 065 (67.0) | 884 (69.0) | 196 (66.7) | 1 498 (67.1) | 598 (69.9) | 49 (61.2) | 2 145 (67.8) |
| Marriage status: n (%) | |||||||
| Married | 617 (38.8) | 459 (35.8) | 113 (38.4) | 838 (37.6) | 317 (37.1) | 34 (42.5) | 1 189 (37.6) |
| Not married | 973 (61.2) | 823 (64.2) | 181 (61.6) | 1 393 (62.4) | 538 (62.9) | 46 (57.5) | 1 977 (62.4) |
| Exercise: n (%) | |||||||
| Current | 457 (28.7) | 358 (27.9) | 84 (28.6) | 653 (29.3) | 229 (26.8) | 17 (21.2) | 899 (28.4) |
| Former | 107 (6.7) | 79 (6.2) | 22 (7.5) | 148 (6.6) | 57 (6.7) | 3 (3.8) | 208 (6.6) |
| Never | 1 024 (64.4) | 845 (65.9) | 188 (63.9) | 1 429 (64.1) | 568 (66.4) | 60 (75.0) | 2 057 (65.0) |
| Missing | 2 (0.1) | 0 (0) | 0 (0) | 1 (0.0) | 1 (0.1) | 0 (0) | 2 (0.1) |
| Smoking: n (%) | |||||||
| Current | 345 (21.7) | 283 (22.1) | 66 (22.4) | 501 (22.5) | 178 (20.8) | 15 (18.8) | 694 (21.9) |
| Former | 207 (13.0) | 176 (13.7) | 34 (11.6) | 264 (11.8) | 139 (16.3) | 14 (17.5) | 417 (13.2) |
| Never | 1 037 (65.2) | 823 (64.2) | 194 (66.0) | 1 465 (65.7) | 538 (62.9) | 51 (63.8) | 2 054 (64.9) |
| Missing | 1 (0.1) | 0 (0) | 0 (0) | 1 (0.0) | 0 (0) | 0 (0) | 1 (0.0) |
| Alcohol drinking: n (%) | |||||||
| Current | 359 (22.6) | 277 (21.6) | 57 (19.4) | 474 (21.2) | 198 (23.2) | 21 (26.2) | 693 (21.9) |
| Former | 176 (11.1) | 118 (9.2) | 23 (7.8) | 216 (9.7) | 91 (10.6) | 10 (12.5) | 317 (10.0) |
| Never | 1 054 (66.3) | 887 (69.2) | 214 (72.8) | 1 540 (69.0) | 566 (66.2) | 49 (61.2) | 2 155 (68.1) |
| Missing | 1 (0.1) | 0 (0) | 0 (0) | 1 (0.0) | 0 (0) | 0 (0) | 1 (0.0) |
| Body mass index group (kg/m 2 ): n (%) | |||||||
| <18 | 487 (30.6) | 392 (30.6) | 104 (35.4) | 689 (30.9) | 269 (31.5) | 25 (31.2) | 983 (31.0) |
| [18, 25) | 924 (58.1) | 725 (56.6) | 165 (56.1) | 1 283 (57.5) | 492 (57.5) | 39 (48.8) | 1 814 (57.3) |
| [25, 30) | 121 (7.6) | 111 (8.7) | 18 (6.1) | 174 (7.8) | 65 (7.6) | 11 (13.8) | 250 (7.9) |
| ≥30 | 22 (1.4) | 27 (2.1) | 3 (1.0) | 37 (1.7) | 13 (1.5) | 2 (2.5) | 52 (1.6) |
| Missing | 36 (2.3) | 27 (2.1) | 4 (1.4) | 48 (2.2) | 16 (1.9) | 3 (3.8) | 67 (2.1) |
Interestingly, the protective effect of FOXO3 is only evident in females. In Table 2, we can see the protective effect of FOXO3 homozygous minor allele carriers, but it became more evident when stratified by gender. The relationship was consistent for all FOXO3 SNPs. Homozygous minor alleles of the 3 FOXO3 SNPs were associated with lower mortality risk in the additive and recessive model. The protective effect tended to be recessive because there was no significant mortality difference between one minor allele and zero minor alleles. The HR (95% CI) adjusted for age and gender in the recessive model were 0.795 (0.668–0.947) for rs4946936, 0.805 (0.689–0.941) for rs2802292, and 0.808 (0.692–0.944) for rs2253310 (Table 2). These associations persisted after adjusting for lifestyle and BMI (data not shown).
Table 2.
The Association Between FOXO3 SNPs and Mortality
| SNP | Minor Allele Number | Total | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | HR (95% CI) | p | n | HR (95% CI) | p | N | HR (95% CI) | p | ||
| rs4946936 | 0 | 1 715 | Reference | — | 788 | Reference | — | 927 | Reference | — |
| rs4946936 | 1 | 1 220 | 0.931 (0.849–1.022) | .13 | 585 | 0.981 (0.854–1.129) | .79 | 635 | 0.897 (0.792–1.016) | .087 |
| rs4946936 | 2 | 231 | 0.771 (0.645–0.922) | .004 | 114 | 0.912 (0.708–1.175) | .48 | 117 | 0.666 (0.517–0.858) | .0016 |
| rs2802292 | 0 | 1 590 | Reference | — | 733 | Reference | — | 857 | Reference | — |
| rs2802292 | 1 | 1 282 | 0.959 (0.874–1.052) | .38 | 602 | 0.96 (0.834–1.105) | .57 | 680 | 0.961 (0.849–1.088) | .53 |
| rs2802292 | 2 | 294 | 0.79 (0.672–0.928) | .004 | 152 | 0.949 (0.758–1.19) | .65 | 142 | 0.667 (0.528–0.843) | .00069 |
| rs2253310 | 0 | 1 609 | Reference | — | 739 | Reference | — | 870 | Reference | — |
| rs2253310 | 1 | 1 261 | 0.946 (0.862–1.039) | .25 | 595 | 0.967 (0.839–1.113) | .64 | 666 | 0.933 (0.824–1.057) | .28 |
| rs2253310 | 2 | 296 | 0.789 (0.671–0.926) | .004 | 153 | 0.948 (0.758–1.187) | .64 | 143 | 0.666 (0.528–0.84) | .00060 |
| rs4946936 | 0 | 1 715 | Reference | — | 788 | Reference | — | 927 | Reference | — |
| rs4946936 | 1/2 | 1 451 | 0.903 (0.826–0.987) | .025 | 699 | 0.969 (0.849–1.107) | .65 | 752 | 0.856 (0.76–0.965) | .011 |
| rs2802292 | 0 | 1 590 | Reference | — | 733 | Reference | — | 857 | Reference | — |
| rs2802292 | 1/2 | 1 576 | 0.924 (0.846–1.01) | .081 | 754 | 0.958 (0.838–1.094) | .52 | 822 | 0.901 (0.8–1.015) | .087 |
| rs2253310 | 0 | 1 609 | Reference | — | 739 | Reference | — | 870 | Reference | — |
| rs2253310 | 1/2 | 1 557 | 0.914 (0.836–0.998) | .045 | 748 | 0.963 (0.843–1.099) | .57 | 809 | 0.879 (0.78–0.99) | .034 |
| rs4946936 | 0/1 | 2 935 | Reference | — | 1 373 | Reference | — | 1 562 | Reference | — |
| rs4946936 | 2 | 231 | 0.795 (0.668–0.947) | .010 | 114 | 0.919 (0.719–1.176) | .50 | 117 | 0.698 (0.545–0.894) | .0044 |
| rs2802292 | 0/1 | 2 872 | Reference | — | 1 335 | Reference | — | 1 537 | Reference | — |
| rs2802292 | 2 | 294 | 0.805 (0.689–0.941) | .0066 | 152 | 0.967 (0.779–1.201) | .76 | 142 | 0.679 (0.542–0.852) | .00084 |
| rs2253310 | 0/1 | 2 870 | Reference | — | 1 334 | Reference | — | 1 536 | Reference | — |
| rs2253310 | 2 | 296 | 0.808 (0.692–0.944) | .0073 | 153 | 0.963 (0.776–1.194) | .73 | 143 | 0.687 (0.548–0.861) | .0011 |
| rs4946936 | 1 | 1 220 | Reference | — | 585 | Reference | — | 635 | Reference | — |
| rs4946936 | 0 | 1 715 | 1.074 (0.978–1.178) | .13 | 788 | 1.019 (0.886–1.172) | .79 | 927 | 1.115 (0.984–1.263) | .087 |
| rs4946936 | 2 | 231 | 0.828 (0.69–0.994) | .042 | 114 | 0.929 (0.717–1.204) | .58 | 117 | 0.743 (0.574–0.961) | .024 |
| rs2802292 | 1 | 1 282 | Reference | — | 602 | Reference | — | 680 | Reference | — |
| rs2802292 | 0 | 1 590 | 1.043 (0.95–1.145) | .38 | 733 | 1.042 (0.905–1.2) | .59 | 857 | 1.041 (0.919–1.178) | .53 |
| rs2802292 | 2 | 294 | 0.824 (0.699–0.971) | .021 | 152 | 0.989 (0.786–1.244) | .93 | 142 | 0.694 (0.548–0.88) | .0025 |
| rs2253310 | 1 | 1 261 | Reference | — | 595 | Reference | — | 666 | Reference | — |
| rs2253310 | 0 | 1 609 | 1.057 (0.963–1.16) | .25 | 739 | 1.035 (0.898–1.191) | .64 | 870 | 1.071 (0.946–1.213) | .28 |
| rs2253310 | 2 | 296 | 0.833 (0.707–0.982) | .029 | 153 | 0.981 (0.78–1.233) | .87 | 143 | 0.714 (0.564–0.904) | .0051 |
Notes: SNP = single-nucleotide polymorphism; HR = hazard ratio; CI = confidence interval. All models adjusted for baseline age and sex.
For SIRT1, the protective effect was seen for some SNPs in the total sample and was only statistically significant for males when stratified by gender (Table 3). Homozygous minor alleles of rs4746720 were associated with lower mortality risk in the recessive (HR [95% CI]: 0.879 [0.782–0.988]) and the heterozygote model (HR [95% CI]: 0.857 [0.757–0.969]), and the results persisted after additionally adjusting for lifestyle and BMI. However, in the context of multiple comparisons, we cannot infer strong associations given chance findings. There was a borderline negative association between the homozygous minor alleles of rs3758391 and mortality risk in the heterozygote model (HR [95% CI]: 0.737 [0.538–1.008]). This association became significant after additionally adjusting for lifestyle and BMI (HR [95% CI]: 0.716 [0.517–0.992]). The association tended to only exist in the male, not in the female (Table 3).
Table 3.
The Association Between SIRT1 SNPs and Mortality
| SNP | Minor Allele Number | Total | Male | Female | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | HR (95% CI) | p | n | HR (95% CI) | p | n | HR (95% CI) | p | ||
| rs12778366 | 0 | 2 334 | Reference | — | 1 090 | Reference | — | 1 244 | Reference | — |
| rs12778366 | 1 | 761 | 1.046 (0.943–1.161) | .39 | 363 | 1.088 (0.934–1.268) | .28 | 398 | 1.008 (0.875–1.162) | .91 |
| rs12778366 | 2 | 71 | 1.071 (0.772–1.486) | .68 | 34 | 1.202 (0.752–1.921) | .44 | 37 | 0.963 (0.609–1.522) | .87 |
| rs3758391 | 0 | 2 231 | Reference | — | 1 041 | Reference | — | 1 190 | Reference | — |
| rs3758391 | 1 | 855 | 1.054 (0.956–1.162) | .29 | 412 | 1.149 (0.993–1.328) | .061 | 443 | 0.987 (0.865–1.127) | .85 |
| rs3758391 | 2 | 80 | 0.776 (0.571–1.056) | .11 | 34 | 0.591 (0.341–1.025) | .061 | 46 | 0.907 (0.626–1.315) | .61 |
| rs2273773 | 0 | 1 693 | Reference | — | 792 | Reference | — | 901 | Reference | — |
| rs2273773 | 1 | 1 246 | 1.083 (0.987–1.188) | .091 | 587 | 1.113 (0.969–1.278) | .13 | 659 | 1.06 (0.936–1.201) | .38 |
| rs2273773 | 2 | 227 | 0.986 (0.827–1.175) | .87 | 108 | 0.929 (0.709–1.216) | .59 | 119 | 1.034 (0.82–1.305) | .78 |
| rs4746720 | 0 | 1 039 | Reference | — | 483 | Reference | — | 556 | Reference | — |
| rs4746720 | 1 | 1 550 | 1.064 (0.963–1.175) | .22 | 754 | 1.012 (0.873–1.173) | .88 | 796 | 1.109 (0.97–1.268) | .13 |
| rs4746720 | 2 | 577 | 0.911 (0.799–1.039) | .16 | 250 | 0.847 (0.69–1.04) | .11 | 327 | 0.959 (0.809–1.138) | .63 |
| rs12778366 | 0 | 2 334 | Reference | — | 1 090 | Reference | — | 1 244 | Reference | — |
| rs12778366 | 1/2 | 832 | 1.048 (0.947–1.16) | .36 | 397 | 1.096 (0.945–1.272) | .23 | 435 | 1.005 (0.875–1.154) | .94 |
| rs3758391 | 0 | 2 231 | Reference | — | 1 041 | Reference | — | 1 190 | Reference | — |
| rs3758391 | 1/2 | 935 | 1.029 (0.935–1.132) | .56 | 446 | 1.1 (0.953–1.268) | .19 | 489 | 0.98 (0.862–1.114) | .76 |
| rs2273773 | 0 | 1 693 | Reference | — | 792 | Reference | — | 901 | Reference | — |
| rs2273773 | 1/2 | 1 473 | 1.067 (0.977–1.166) | .15 | 695 | 1.082 (0.948–1.236) | .24 | 778 | 1.056 (0.938–1.189) | .37 |
| rs4746720 | 0 | 1 039 | Reference | — | 483 | Reference | — | 556 | Reference | — |
| rs4746720 | 1/2 | 2 127 | 1.019 (0.928–1.12) | .69 | 1 004 | 0.969 (0.841–1.115) | .66 | 1 123 | 1.062 (0.936–1.204) | .35 |
| rs12778366 | 0/1 | 3 095 | Reference | — | 1 453 | Reference | — | 1 642 | Reference | — |
| rs12778366 | 2 | 71 | 1.059 (0.764–1.468) | .73 | 34 | 1.176 (0.737–1.877) | .50 | 37 | 0.961 (0.609–1.516) | .86 |
| rs3758391 | 0/1 | 3 086 | Reference | — | 1 453 | Reference | — | 1 633 | Reference | — |
| rs3758391 | 2 | 80 | 0.765 (0.563–1.038) | .085 | 34 | 0.568 (0.328–0.983) | .043 | 46 | 0.911 (0.63–1.318) | .62 |
| rs2273773 | 0/1 | 2 939 | Reference | — | 1 379 | Reference | — | 1 560 | Reference | — |
| rs2273773 | 2 | 227 | 0.953 (0.803–1.131) | .58 | 108 | 0.887 (0.682–1.153) | .37 | 119 | 1.009 (0.805–1.266) | .93 |
| rs4746720 | 0/1 | 2 589 | Reference | — | 1 237 | Reference | — | 1 352 | Reference | — |
| rs4746720 | 2 | 577 | 0.879 (0.782–0.988) | .030 | 250 | 0.841 (0.699–1.011) | .066 | 327 | 0.904 (0.777–1.051) | .19 |
| rs12778366 | 1 | 761 | Reference | — | 363 | Reference | — | 398 | Reference | — |
| rs12778366 | 0 | 2 334 | 0.956 (0.861–1.06) | .39 | 1 090 | 0.919 (0.788–1.071) | .28 | 1 244 | 0.992 (0.861–1.143) | .91 |
| rs12778366 | 2 | 71 | 1.023 (0.732–1.431) | .89 | 34 | 1.104 (0.683–1.786) | .69 | 37 | 0.955 (0.598–1.526) | .85 |
| rs3758391 | 1 | 855 | Reference | — | 412 | Reference | — | 443 | Reference | — |
| rs3758391 | 0 | 2 231 | 0.949 (0.86–1.046) | .29 | 1 041 | 0.871 (0.753–1.007) | .061 | 1 190 | 1.013 (0.887–1.157) | .85 |
| rs3758391 | 2 | 80 | 0.737 (0.538–1.008) | .056 | 34 | 0.515 (0.295–0.899) | .020 | 46 | 0.919 (0.628–1.345) | .66 |
| rs2273773 | 1 | 1 246 | Reference | — | 587 | Reference | — | 659 | Reference | — |
| rs2273773 | 0 | 1 693 | 0.923 (0.842–1.013) | .091 | 792 | 0.899 (0.783–1.032) | .13 | 901 | 0.943 (0.833–1.068) | .36 |
| rs2273773 | 2 | 227 | 0.91 (0.761–1.089) | .30 | 108 | 0.835 (0.635–1.097) | .19 | 119 | 0.976 (0.769–1.237) | .84 |
| rs4746720 | 1 | 1 550 | Reference | — | 754 | Reference | — | 796 | Reference | — |
| rs4746720 | 0 | 1 039 | 0.94 (0.851–1.038) | .22 | 483 | 0.988 (0.853–1.146) | .88 | 556 | 0.902 (0.789–1.031) | .13 |
| rs4746720 | 2 | 577 | 0.857 (0.757–0.969) | .014 | 250 | 0.837 (0.69–1.016) | .072 | 327 | 0.865 (0.737–1.016) | .078 |
Notes: SNP = single-nucleotide polymorphism; HR = hazard ratio; CI = confidence interval. All models adjusted for baseline age and sex.
We identified significant interactions between rs4946936 and rs12778366, rs4946936 and rs3758391, rs2802292 and rs3758391, and rs2253310 and rs2273773 (Table 4). In the stratified analyses, homozygous minor alleles of rs2802292 had lower mortality risk compared to homozygous major alleles for participants carrying homozygous major alleles of rs3758391 (Supplementary Table 3). Homozygous minor alleles of rs3758391 had lower mortality risk than homozygous major alleles for participants carrying homozygous major alleles of rs2802292 (Supplementary Table 4).
Table 4.
Significant Interaction of SIRT1 and FOXO SNPs on Mortality
| Without Interaction | With Interaction | ||||
|---|---|---|---|---|---|
| Minor Allele Number (0 as reference) | HR (95% CI) | p | HR (95% CI) | Beta (SE) | p |
| rs4946936 one | 0.931 (0.849–1.022) | .13 | 0.917 (0.823–1.021) | −0.087 (0.055) | .11 |
| rs4946936 two | 0.771 (0.645–0.922) | .0044 | 0.739 (0.599–0.912) | −0.303 (0.107) | .0048 |
| rs12778366 one | 1.047 (0.944–1.162) | .38 | 1.022 (0.886–1.178) | 0.021 (0.073) | .78 |
| rs12778366 two | 1.058 (0.763–1.469) | .73 | 0.907 (0.588–1.4) | −0.097 (0.221) | .66 |
| rs4946936 one × rs12778366 one | — | — | 1.049 (0.844–1.305) | 0.048 (0.111) | .66 |
| rs4946936 two × rs12778366 one | — | — | 1.087 (0.717–1.648) | 0.083 (0.212) | .70 |
| rs4946936 one × rs12778366 two | — | — | 1.257 (0.614–2.577) | 0.229 (0.366) | .53 |
| rs4946936 two × rs12778366 two | — | — | 3.199 (1.076–9.517) | 1.163 (0.556) | .037 |
| rs4946936 one | 0.929 (0.847–1.02) | .12 | 0.817 (0.73–0.915) | −0.202 (0.058) | .00047 |
| rs4946936 two | 0.769 (0.643–0.919) | .0039 | 0.748 (0.603–0.927) | −0.291 (0.11) | .0080 |
| rs3758391 one | 1.053 (0.955–1.161) | .30 | 0.894 (0.78–1.025) | −0.112 (0.07) | .11 |
| rs3758391 two | 0.767 (0.564–1.043) | .090 | 0.623 (0.415–0.935) | −0.474 (0.207) | .022 |
| rs4946936 one × rs3758391 one | — | — | 1.483 (1.208–1.82) | 0.394 (0.105) | .00016 |
| rs4946936 two × rs3758391 one | — | — | 1.094 (0.737–1.622) | 0.089 (0.201) | .66 |
| rs4946936 one × rs3758391 two | — | — | 1.841 (0.968–3.502) | 0.61 (0.328) | .063 |
| rs4946936 two × rs3758391 two | — | — | 1.014 (0.236–4.362) | 0.014 (0.744) | .98 |
| rs2802292 one | 0.959 (0.874–1.052) | .38 | 0.852 (0.761–0.954) | −0.16 (0.058) | .0055 |
| rs2802292 two | 0.787 (0.67–0.926) | .0038 | 0.741 (0.609–0.902) | −0.299 (0.1) | .0028 |
| rs3758391 one | 1.054 (0.955–1.162) | .29 | 0.888 (0.77–1.024) | −0.119 (0.073) | .10 |
| rs3758391 two | 0.77 (0.566–1.047) | .095 | 0.621 (0.398–0.969) | −0.476 (0.227) | .036 |
| rs2802292 one × rs3758391 one | — | — | 1.447 (1.177–1.778) | 0.369 (0.105) | .00044 |
| rs2802292 two × rs3758391 one | — | — | 1.199 (0.841–1.709) | 0.182 (0.181) | .31 |
| rs2802292 one × rs3758391 two | — | — | 1.576 (0.832–2.984) | 0.455 (0.326) | .16 |
| rs2802292 two × rs3758391 two | — | — | 1.464 (0.428–5.005) | 0.381 (0.627) | .54 |
| rs2253310 one | 0.947 (0.863–1.04) | .25 | 0.832 (0.743–0.932) | −0.184 (0.058) | .0015 |
| rs2253310 two | 0.787 (0.67–0.924) | .0035 | 0.727 (0.597–0.884) | −0.319 (0.1) | .0014 |
| rs3758391 one | 1.052 (0.954–1.161) | .31 | 0.874 (0.759–1.006) | −0.135 (0.072) | .060 |
| rs3758391 two | 0.769 (0.566–1.046) | .094 | 0.605 (0.388–0.944) | −0.502 (0.227) | .027 |
| rs2253310 one × rs3758391 one | — | — | 1.5 (1.22–1.845) | 0.406 (0.105) | .00012 |
| rs2253310 two × rs3758391 one | — | — | 1.269 (0.894–1.801) | 0.238 (0.179) | .18 |
| rs2253310 one × rs3758391 two | — | — | 1.673 (0.883–3.168) | 0.514 (0.326) | .11 |
| rs2253310 two × rs3758391 two | — | — | 1.514 (0.443–5.179) | 0.415 (0.627) | .51 |
| rs2253310 one | 0.943 (0.858–1.035) | .21 | 1.051 (0.925–1.195) | 0.05 (0.065) | .45 |
| rs2253310 two | 0.788 (0.671–0.925) | .0036 | 0.771 (0.613–0.968) | −0.261 (0.116) | .025 |
| rs2273773 one | 1.086 (0.99–1.192) | .080 | 1.178 (1.034–1.342) | 0.164 (0.066) | .014 |
| rs2273773 two | 0.991 (0.831–1.182) | .92 | 1.133 (0.887–1.447) | 0.125 (0.125) | .32 |
| rs2253310 one × rs2273773 one | — | — | 0.81 (0.666–0.984) | −0.211 (0.1) | .034 |
| rs2253310 two × rs2273773 one | — | — | 1.049 (0.749–1.469) | 0.048 (0.172) | .78 |
| rs2253310 one × rs2273773 two | — | — | 0.72 (0.494–1.049) | −0.329 (0.192) | .087 |
| rs2253310 two × rs2273773 two | — | — | 0.98 (0.537–1.787) | −0.02 (0.307) | .95 |
Notes: SNP = single-nucleotide polymorphism; HR = hazard ratio; CI = confidence interval. All models adjusted for baseline age and sex.
There was no significant 3-way interaction of FOXO3, SIRT1, and gender (Supplementary Table 5).
The protective effect of FOXO3 and SIRT1 SNPs was also illustrated in the adjusted survival curve shown in Supplementary Figures 1 and 2. After combining the genotype of rs2802292 and rs3758391, those with at least one minor allele of rs2802292 and without minor allele of rs3758391 (blue line), and those with at least one minor allele of rs3758391 and without minor allele of rs2802292 (green line) had higher survival rate than those without any minor allele of rs2802292 and rs3758391 (red line), and those with at least one minor allele of both rs2802292 and rs3758391 (purple line; Supplementary Figure 3).
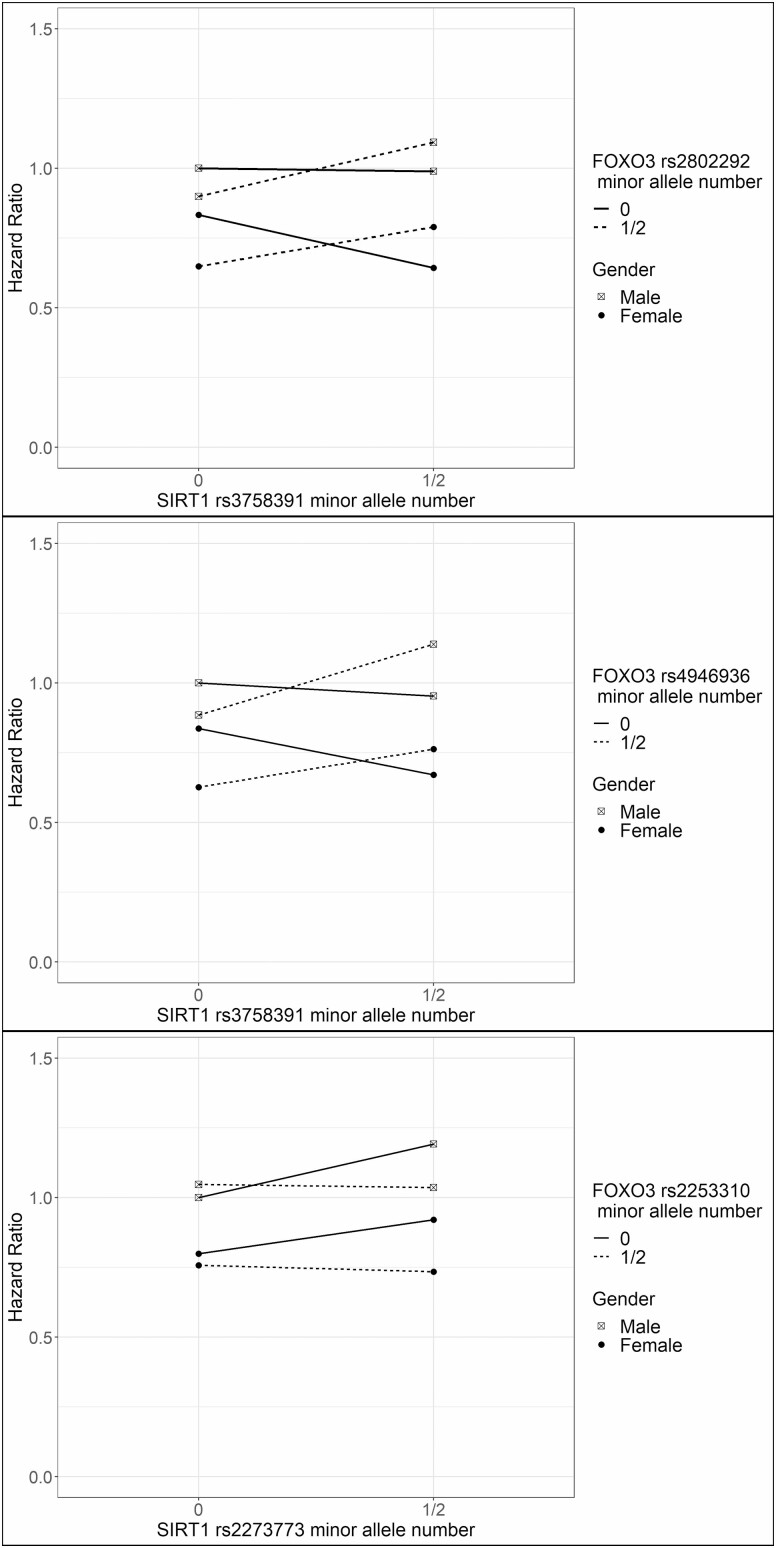
In Figure 1 and Supplementary Figure 4, rs2802292 minor allele carriers showed lower mortality risk than rs2802292 minor allele noncarriers in both male and female rs3758391 minor allele noncarriers, but this association became reversed in both male and female rs3758391 minor allele carriers. Meanwhile, the association was stronger in females than males. rs3758391 minor allele carriers showed lower mortality risk than rs3758391 minor allele noncarriers in females rs2802292 minor allele noncarriers, but this association became reversed in both male and female rs2802292 minor allele carriers. Females had a lower mortality risk than males in all genotype combinations.
Figure 1.
The hazard ratios of different combinations of FOXO3, SIRT1 SNPs, and gender. Note: The model adjusted for age at baseline. SNP = single-nucleotide polymorphism.
Discussion
In this cohort study with up to 10 years of follow-up, we found evidence of a strong protective effect of FOXO3 against mortality across all the studied SNPs. The effect was only evident in female study participants. For SIRT1, we found some but not strong protective effects against mortality, with the effect only evident for male study participants. Furthermore, we found an interactive effect of FOXO3 and SIRT1. However, the effect was not synergistic. When FOXO3 and SIRT1 genes co-occur, it appears that the FOXO3 effect is more dominant. Female populations in our study appear to have the most mortality benefit from FOXO3.
In 2008, a strong association between FOXO3 and human longevity was firstly reported by Willcox et al. (10) in a long-lived population of male Americans of Japanese and Okinawan ancestry. This novel finding was then replicated in centenarians of German ancestry (11), followed by other populations, including Caucasian women (12), Italians (13), Chinese (7), and others. Sex difference in genetic determinants of longevity has been suggested to be overlooked in GWAS and observational cohorts (6) because investigators typically do not assume there are gender differences in genetic effects. While women typically live longer than men globally, current studies could not quantify whether this is due to the genetic advantage of women or harmful lifestyle (smoking, drinking) and occupational exposures (job hazard, environmental chemical exposure) of men (14,15). Previous studies suggested that the estrogen level may affect the FOXO3 regulatory region and lead to sex differences in aging (16). However, given the advanced age of our female study participants, which are likely to be postmenopausal, we are not sure if this mechanism holds. In animal models, FOXO3 phosphorylation was lower in females (17). Phosphorylation represents a reversible mechanism employed by cells to regulate transcription factor activity in response to alterations in the extracellular environment. We lack evidence on whether molecular pathways (transcription factors, transcriptional coregulators) of phosphorylation protein kinases or dephosphorylation by protein phosphatases is different by sex. An earlier study using the same CLHLS cohort found FOXO3 was associated with expanded life span in both genders (7). However, the study used a case–control study design without the longitudinal follow-up data in our study. Another CLHLS study used polygenic risk score analyses to examine sex differences and found the pathway of sex-specific loci and longevity different between sexes (6). A review article summarizing cohort findings of FOXO3 on longevity in the cohort of American men of Japanese ancestry along with 11 independent studies of populations of diverse ancestry in multiple different countries did not mention sex differences, possibly due to study design constraints (18). Newer disease-specific studies show that FOXO3 has been shown to increase the life span of at-risk individuals (men) by protection against cardiometabolic stress (19). Women express higher levels of FOXO3 mRNA than men in skeletal muscle tissues (20). While our study does not challenge the association of FOXO3 on longevity in both genders (21), our findings indicate that the contribution may be unequal.
Literature for SIRT1 and sex differences in the human population also point to the role of estrogen as a modifying factor. A study showed that SIRT1 protects arteries against menopause-induced senescence and atherosclerosis (22). Another study showed female sex-specific downregulation of SIRT1 in aged hearts (23). Another study points to age and sex modifications of SIRT1, with women in their 30s showing the highest SIRT1 activities (24). The sex differences in the roles of SIRT1 in many disease outcomes remain to be explored further. No study has presented robust findings on mortality outcomes with respect to gender differences.
As both FOXO3 and SIRT1 are recognized longevity genes, prior research assessed how they might work together. In the worm model, NAD+-dependent SIRT1 extends life span by utilizing the FOXO transcription factor daf-16. Several findings focusing on mammalian SIRT1 and FOXO have highlighted this genetic interaction. It is hypothesized that mammalian SIRT1 deacetylates FOXO and reduces apoptosis and potentiating FOXO-induced cell cycle arrest. SIRT1 might increase longevity by shifting FOXO-dependent responses away from cell death (19). Other studies point to mechanisms involving bone loss by vitamin D deficiency (25) and may be activated under oxidative stress (26). Likely, the mechanism is multifaceted. The combined and synergistic effect of FOXO3 and SIRT1 has not been studied to date in population cohorts. Compared to those only carrying FOXO3 protective alleles, our study did not see the advantages of carrying both FOXO3 and SIRT1 protective alleles with respect to all-cause mortality.
Our study has several notable strengths. First, using an observational study with a high proportion of the oldest old, we can see real-world evidence of the genetic benefit of FOXO3 and SIRT1. This cohort of older individuals also resides in China, which provides generalizability evidence to add to the findings from prior cohorts. Second, our cohort is a longitudinal study with over a decade of follow-up, which means we can capture more mortality events. Third, our cohort is rich in characteristic demographic information, which allows us to conduct stratified analyses and adjust for potential confounders. Limitations of our study findings include the lack of cause-specific mortality information, which would inform which disease outcome SIRT1 and FOXO3 may prevent. Our study lacks molecular and epigenetic markers, which does not allow us to see in detail which biological pathway is upregulated or downregulated by genes.
In conclusion, we found strong gene-by-gender interaction. Strong FOXO3 effects were driven by female study participants, and protective effects of the SIRT1 effect were in male study participants. There does not appear to be a synergistic effect of being carriers of both longevity candidate gene SNPs. Overall, our findings add novel information that female and male populations may have different benefits from SIRT1 and FOXO3 genetic SNPs. Whether this is due to gene–environmental interaction or hormonal differences remains unexplored.
Supplementary Material
Acknowledgments
The authors extend appreciation to the participants and investigators of the Chinese Longitudinal Healthy Longevity Survey study.
Contributor Information
John S Ji, Vanke School of Public Health, Tsinghua University, Beijing, China.
Linxin Liu, Vanke School of Public Health, Tsinghua University, Beijing, China.
Chang Shu, Departments of Pediatrics and Systems Biology, Columbia University, New York, New York, USA.
Lijing L Yan, Global Health Research Center, Duke Kunshan University, Kunshan, China.
Yi Zeng, Center for Healthy Aging and Development Studies, National School of Development, Peking University, Beijing, China; Center for the Study of Aging and Human Development, Duke Medical School, Durham, North Carolina, USA.
Funding
The Chinese Longitudinal Healthy Longevity Study (CLHLS) data sets analyzed in this article are jointly supported by the National Key R&D Program of China (2018YFC2000400), National Natural Sciences Foundation of China (72061137004, 71490732) the U.S. National Institute of Aging of National Institute of Health (P01AG031719), and Duke University School of Medicine and Duke-NUS Medical School/RECA (Pilot)/2019/0051.
Conflict of Interest
None declared.
References
- 1. Brunet A, Sweeney LB, Sturgill JF, et al. . Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637 [DOI] [PubMed] [Google Scholar]
- 2. Kobayashi Y, Furukawa-Hibi Y, Chen C, et al. . SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16(2):237–243. doi: 10.3892/ijmm.16.2.237 [DOI] [PubMed] [Google Scholar]
- 3. Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14(8): 408–412. doi: 10.1016/j.tcb.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 4. Oellerich MF, Potente M. FOXOs and sirtuins in vascular growth, maintenance, and aging. Circ Res. 2012;110(9):1238–1251. doi: 10.1161/CIRCRESAHA.111.246488 [DOI] [PubMed] [Google Scholar]
- 5. Kedenko L, Lamina C, Kedenko I, et al. . Genetic polymorphisms at SIRT1 and FOXO1 are associated with carotid atherosclerosis in the SAPHIR cohort. BMC Med Genet. 2014;15:112. doi: 10.1186/s12881-014-0112-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zeng Y, Nie C, Min J, et al. . Sex differences in genetic associations with longevity. JAMA Netw Open. 2018;1(4):e181670. doi: 10.1001/jamanetworkopen.2018.1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Y, Wang WJ, Cao H, et al. . Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18(24):4897–4904. doi: 10.1093/hmg/ddp459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yao Y, Liu L, Guo G, Zeng Y, Ji JS. Interaction of Sirtuin 1 (SIRT1) candidate longevity gene and particulate matter (PM2.5) on all-cause mortality: a longitudinal cohort study in China. Environ Health. 2021;20(1):25. doi: 10.1186/s12940-021-00718-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Therneau TM, Crowson CS, Atkinson Jan EJ. Adjusted survival curves. 2015. https://cran.r-project.org/web/packages/survival/vignettes/adjcurve.pdf. Accessed October 1, 2021.
- 10. Willcox BJ, Donlon TA, He Q, et al. . FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105(37): 13987–13992. doi: 10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flachsbart F, Caliebe A, Kleindorp R, et al. . Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106(8):2700–2705. doi: 10.1073/pnas.0809594106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pawlikowska L, Hu D, Huntsman S, et al. ; Study of Osteoporotic Fractures . Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8(4):460–472. doi: 10.1111/j.1474-9726.2009.00493.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Anselmi CV, Malovini A, Roncarati R, et al. . Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12(2):95–104. doi: 10.1089/rej.2008.0827 [DOI] [PubMed] [Google Scholar]
- 14. Zarulli V, Barthold Jones JA, Oksuzyan A, Lindahl-Jacobsen R, Christensen K, Vaupel JW. Women live longer than men even during severe famines and epidemics. Proc Natl Acad Sci U S A. 2018;115(4):E832–E840. doi: 10.1073/pnas.1701535115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baum F, Musolino C, Gesesew HA, Popay J. New perspective on why women live longer than men: an exploration of power, gender, social determinants, and capitals. Int J Environ Res Public Health. 2021;18(2):661. doi: 10.3390/ijerph18020661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanese P, Forte G, Disciglio V, Grossi V, Simone C. FOXO3 on the road to longevity: lessons from SNPs and chromatin hubs. Comput Struct Biotechnol J. 2019;17:737–745. doi: 10.1016/j.csbj.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshihara T, Natsume T, Tsuzuki T, et al. . Sex differences in forkhead box O3a signaling response to hindlimb unloading in rat soleus muscle. J Physiol Sci. 2019;69(2):235–244. doi: 10.1007/s12576-018-0640-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morris BJ, Willcox DC, Donlon TA,Willcox BJ.. FOXO3: a major gene for human longevity - a mini-review. Gerontology. 2015;61(6):515–525. doi: 10.1159/000375235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen R, Morris BJ, Donlon TA, et al. . FOXO3 longevity genotype mitigates the increased mortality risk in men with a cardiometabolic disease. Aging (Albany NY). 2020;12(23):23509–23524. doi: 10.18632/aging.202175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welle S, Tawil R, Thornton CA. Sex-related differences in gene expression in human skeletal muscle. PLoS One. 2008;3(1):e1385. doi: 10.1371/journal.pone.0001385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flachsbart F, Caliebe A, Kleindorp R, et al. . Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106(8):2700–2705. doi: 10.1073/pnas.0809594106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasaki Y, Ikeda Y, Miyauchi T, Uchikado Y, Akasaki Y, Ohishi M. Estrogen–SIRT1 axis plays a pivotal role in protecting arteries against menopause-induced senescence and atherosclerosis. J Atheroscler Thromb. 2020;27(1):47–59. doi: 10.5551/jat.47993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barcena de Arellano ML, Pozdniakova S, Kühl AA, Baczko I, Ladilov Y, Regitz-Zagrosek V. Sex differences in the aging human heart: decreased sirtuins, pro-inflammatory shift and reduced anti-oxidative defense. Aging (Albany NY). 2019;11(7):1918–1933. doi: 10.18632/aging.101881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee HJ, Yang SJ. Aging-related correlation between serum Sirtuin 1 activities and basal metabolic rate in women, but not in men. Clin Nutr Res. 2017;6(1):18–26. doi: 10.7762/cnr.2017.6.1.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen H, Hu X, Yang R, et al. . SIRT1/FOXO3a axis plays an important role in the prevention of mandibular bone loss induced by 1,25(OH)2D deficiency. Int J Biol Sci. 2020;16(14):2712–2726. doi: 10.7150/ijbs.48169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hori YS, Kuno A, Hosoda R, Horio Y. Regulation of FOXOs and p53 by SIRT1 modulators under oxidative stress. PLoS One. 2013;8(9):e73875. doi: 10.1371/journal.pone.0073875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.