Extended Data Fig. 9. Meningioma DNA methylation grouping schemes uncontrolled for the influence of CNVs on β values.
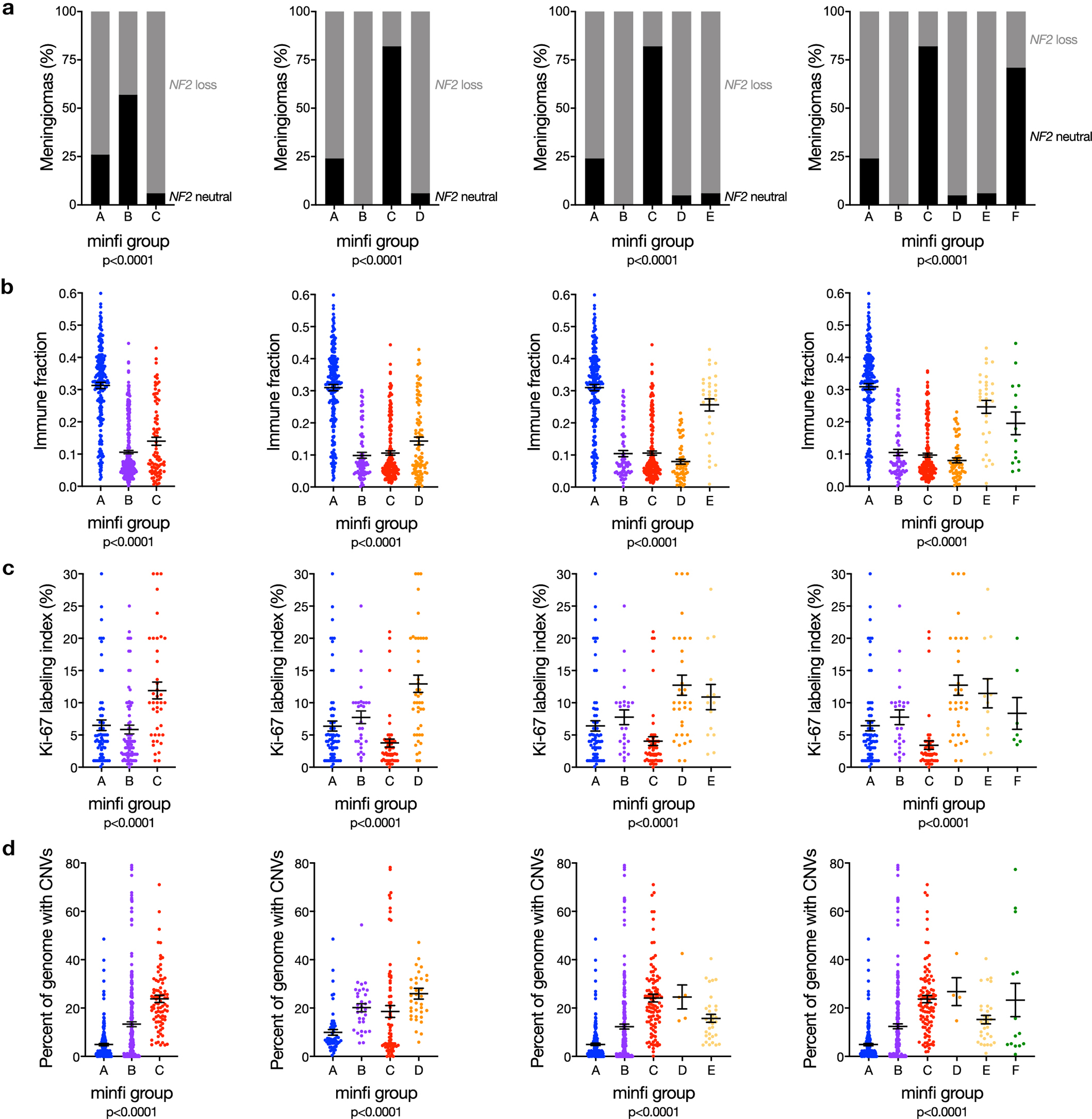
a, Meningioma DNA methylation analysis of copy number loss at the NF2 locus (n=565) across different numbers of DNA methylation groups determined by the minfi pre-processing pipeline (Chi-squared tests, two-sided). b, Meningioma DNA methylation estimation of leukocyte fraction (n=565) across different numbers of DNA methylation groups determined by the minfi pre-processing pipeline (ANOVA, one-sided). c, Ki-67 labeling index from meningioma clinical pathology reports (n=206) across different numbers of DNA methylation groups determined by the minfi pre-processing pipeline (ANOVA, one-sided). b, Meningioma genomes (n=565) with copy number variations (CNVs) across DNA methylation groups determined by the minfi pre-processing pipeline (ANOVA, one-sided). Regardless of the number of groups, meningioma DNA methylation analysis uncontrolled for the influence of CNVs on β values cannot identify a grouping scheme with non-redundant differences in clinical outcomes (Extended data figure 3d), NF2 loss, immune enrichment, cell proliferation, and chromosome instability. Lines represent means, and error bars represent standard error of the means. minfi meningioma DNA methylation grouping schemed comprised of 3, 4, 5, or 6 groups are designated by letters A-C, A-D, A-E, or A-F, respectively.
