Extended Data Fig. 10. Immune-enriched meningiomas display markers of T cell exhaustion and immunoediting.
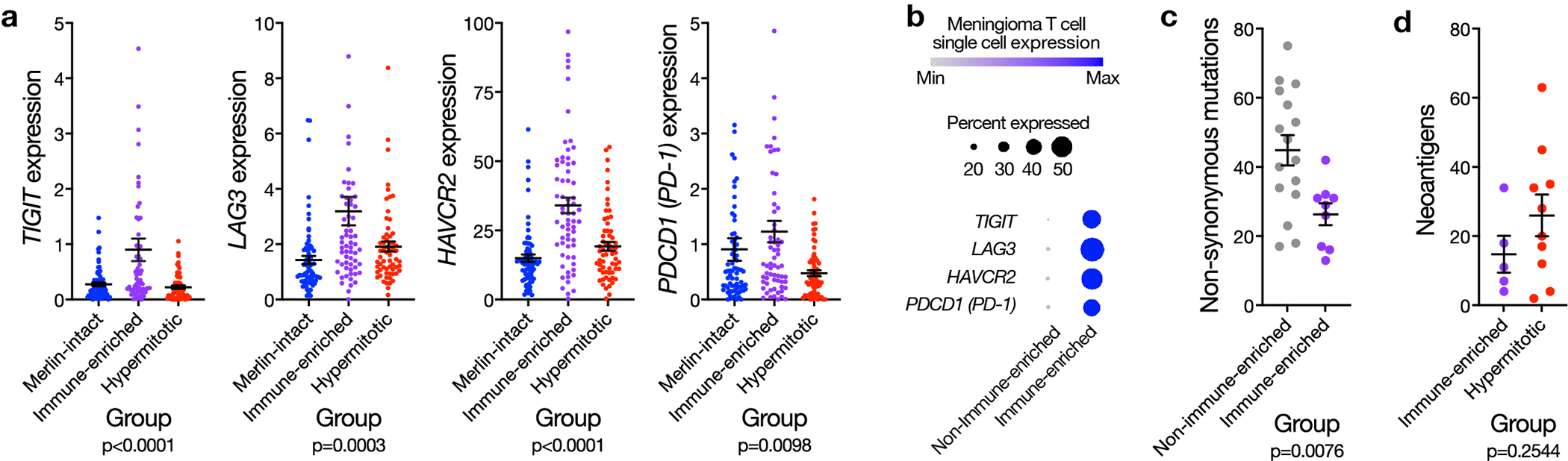
a, Meningioma transcripts per million (TPM) expression of TIGIT, LAG3, HAVCR2, or PDCD1 (n=200) T cell exhaustion markers across DNA methylation groups. Lines represent means, and error bars represent standard error of the means (ANOVA, one-sided). b, Single-cell RNA sequencing relative expression of immune exhaustion genes in T cells across Immune-enriched (n=5) and non-Immune-enriched (n=3) meningioma samples. Circle size denotes percentage of cells. Circle shading denotes average expression. c, Non-synonymous mutations from whole-exome sequencing of Immune-enriched (n=9) and non-Immune-enriched (n=16) and meningiomas, with paired normal samples, overlapping with the discovery cohort. Lines represent means, and error bars represent standard error of the means (Student’s t test, one-sided). d, Neoantigen prediction from whole-exome sequencing of Immune-enriched (n=5) and Hypermitotic (n=9) meningiomas, with paired normal samples, overlapping with the discovery cohort. Lines represent means, and error bars represent standard error of the means (Student’s t test, one-sided).
