Figure 6. Genetic, epigenetic, and transcriptomic mechanisms distinguishing meningioma DNA methylation groups.
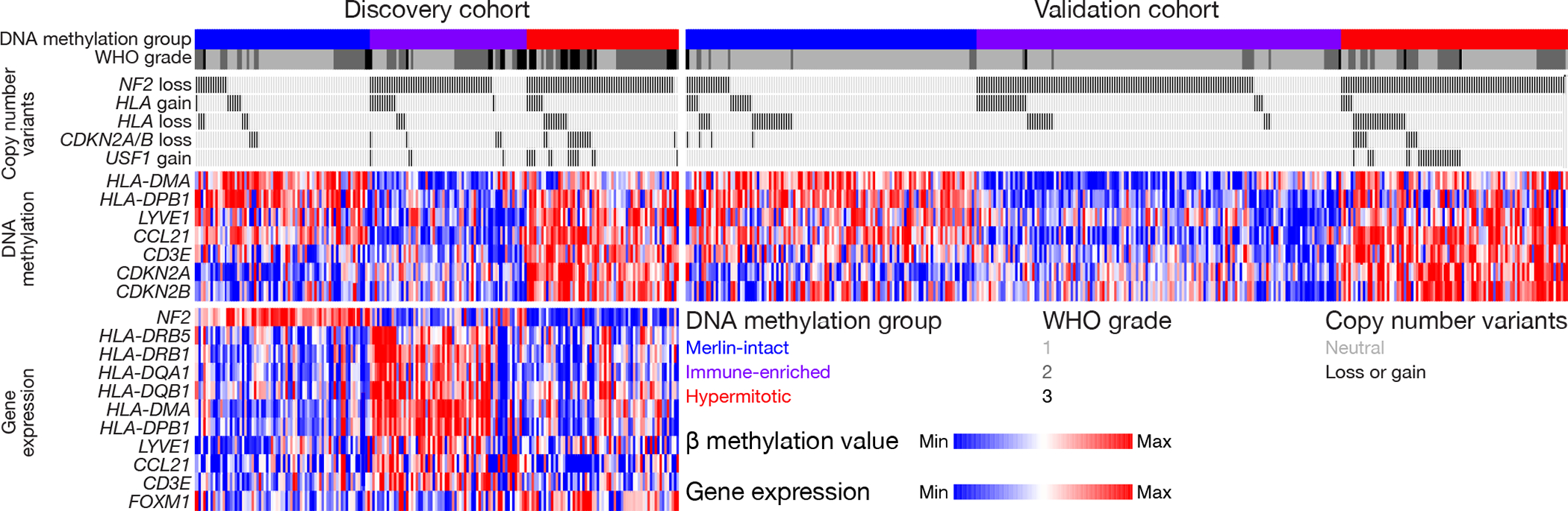
Oncoprint comprised of the 565 meningiomas in this study. CNVs of any size deleting or amplifying entire genes, scaled β methylation values, and scaled transcripts per million (TPM) are shown. The focal versus broad nature of CNVs are described in the Copy number analysis section of the Methods. HLA gain/loss shows the polymorphic locus encompassing HLA-DRB5, HLA-DRB1, HLA-DQA1, and HLA-DQB1. β methylation values and TPM are scaled from the bottom 10th percentile to the top 90th percentile of each row. RNA sequencing was performed on the discovery cohort (n=200) but not on the validation cohort.
