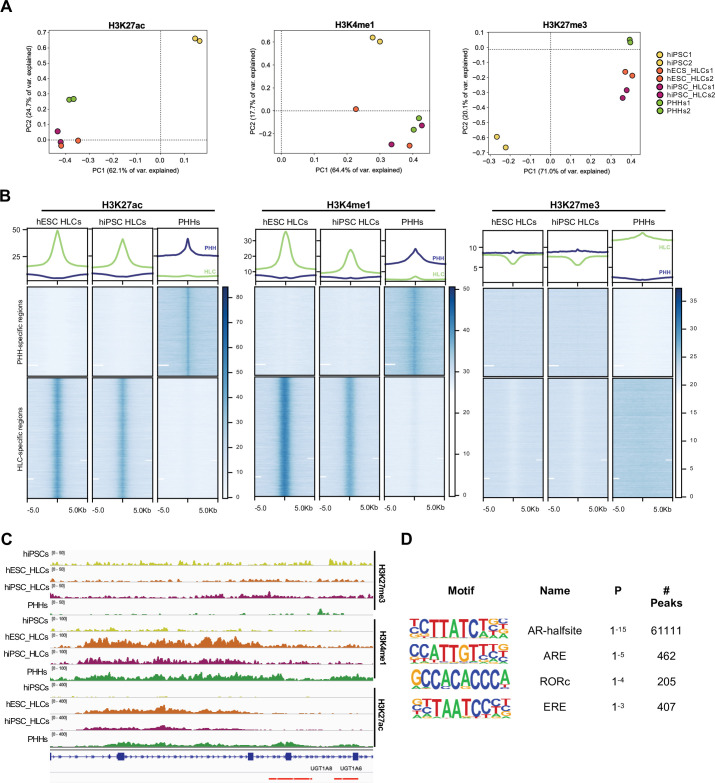
Figure 3. Epigenetic status of regulatory regions differs between states of maturation in hepatocyte-like cells (HLCs) and primary human hepatocytes (PHHs).
(A) Principal component analysis (PCA) of the global enrichment profile of H3K27ac, H3K4me1, and H3K27me3 across two replicates of undifferentiated human induced pluripotent stem cells (hiPSCs), human embryonic stem cell (hESC), and hiPSC-derived HLCs, and PHHs. Average scores were computed for genomic regions of 1000 bp for the entire genome. (B) Average density plots and heatmaps showing enrichment levels for H3K27ac, H3K4me1, and H3K27me3 within a 10 kb window centred at H3K27ac PHH-unique (blue) or HLC-unique (green) regions. Scales are adjusted to maximum peak intensity for each dataset. (C) Enrichment profiles of H3K27ac, H3K4me1, and H3K27me3 across the UGT1A locus. Profiles are shown for one replicate of undifferentiated hiPSCs, hESC, and hiPSC-derived HLCs, and PHHs. Red bars represent PHH-unique H3K27ac peaks. (D) Nuclear receptor motifs identified as overrepresented binding sites at H3K27ac PHH-unique regions.