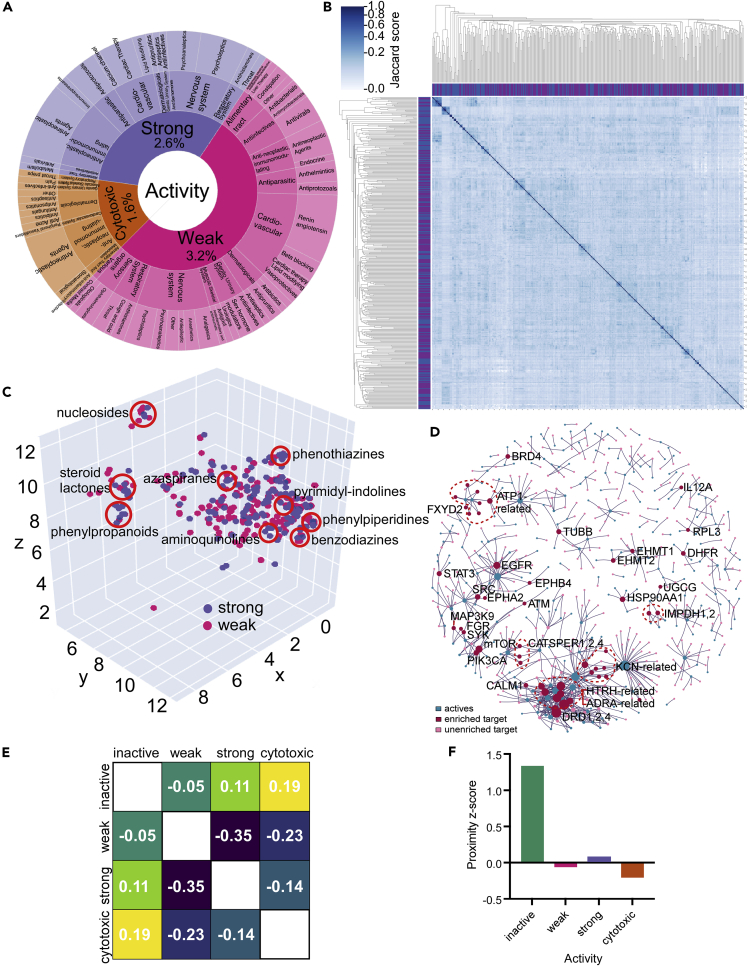
Figure 2.
Computational analysis of primary screen outcomes
(A) Chart showing the active compounds by ATC classifiers for strongly active, weakly active, and cytotoxic compounds. The fraction of compounds in each category is shown as a percentage of all compounds in the library.
(B) Pairwise comparison of active compounds based on computed structural similarity using Morgan fingerprints and Jaccard analysis.
(C) 3D representation of structural similarity of active molecules based on the reduced dimensionality of the molecular bit vectors. Localized clusters of major structural classes are indicated by red circles and are approximate locations in the plot. Refer to Figure S3 for an interactive version with higher detail.
(D) Relationship of enriched protein drug targets (red circles) or unenriched targets (pink circles) with active compounds (blue circles) for compounds with annotated targets in the DRH database.
(E) Separation heatmap between the targets of different activity classes within the human protein–protein interaction network. Negative network separation values reflect overlapping neighborhoods.
(F) Proximity Z-score between the drug targets of each category and the SARS-CoV-2 protein-binding host factors in the human protein-protein interaction network. Drugs with treatment activity have Z-scores close to zero, much lower than inactive drugs, indicating they hit targets in the network proximity of SARS-CoV-2-binding proteins.