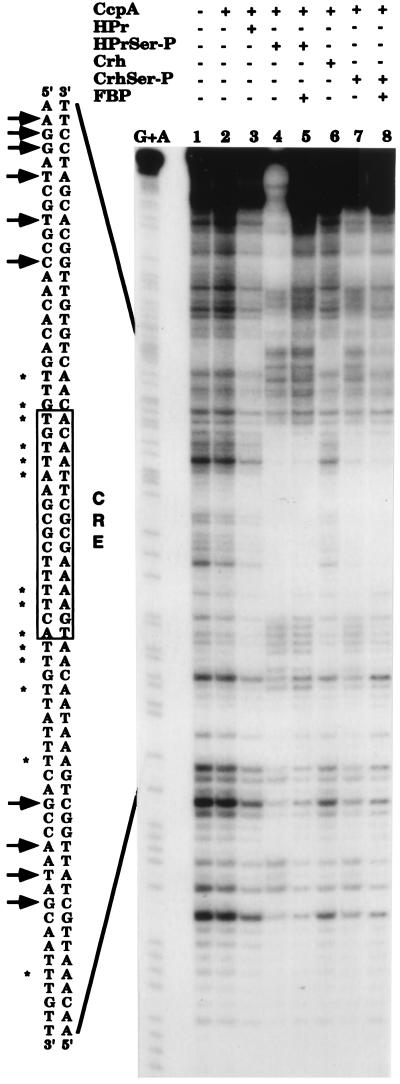
FIG. 3.
DNase I footprinting experiments with the lev cre sequence in the presence of CcpA, P-Ser–HPr, and P-Ser–Crh. A 178-bp EcoRI-PstI fragment (from positions −148 to +30) containing the lev promoter and the cre sequence (from position −50 to −36) was labeled at the 3′-end as described in the text. DNA was digested with DNase I in the absence of any protein (lane 1) or in the presence of 5 μM CcpA (lane 2); 2.5 μM CcpA and 10 μM HPr (lane 3), 2.5 μM CcpA and 10 μM P-Ser–HPr (lane 4), 2.5 μM CcpA, 10 μM P-Ser–HPr, and 20 mM FBP (lane 5), 2.5 μM CcpA and 10 μM Crh (lane 6), 2.5 μM CcpA and 10 μM P-Ser–Crh (lane 7), or 2.5 μM CcpA, 10 μM P-Ser–Crh, and 20 mM FBP (lane 8). The base-specific chemical cleavage reaction at guanine and adenine (20) is shown in lane G+A. The cre sequence is boxed, and the cre consensus sequence is indicated. The bases protected against digestion by DNase I are indicated by asterisks, while the sites of hyperdigestion by DNase I are indicated by arrows.