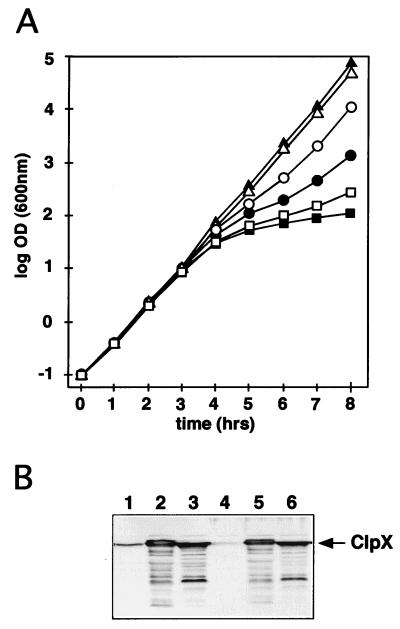
FIG. 3.
clpXCc is able to complement an E. coli clpX mutant. (A) The activity of ClpX was monitored by its ability to degrade, together with ClpP, the antidote protein Phd of the P1 plasmid addiction module Phd-Doc (26). Loss of the phd gene results in Doc-dependent cell killing if the Phd protein is degraded by the ClpXP protease. Growth of cultures containing the phd and doc genes on a plasmid with a temperature-sensitive replicon (pGB2ts::phd-doc) was monitored after a shift to the nonpermissive temperature. Growth is shown as the log OD600. The time after the temperature increase is indicated in hours. Cessation of growth 3 to 5 h after the temperature shift was an indicator of the rapid disappearance of the antidote protein Phd and thus of ClpX activity. The following plasmids were used: plasmid pGB2ts is temperature sensitive for replication; pGB2ts::phd-doc is identical to pGB2ts except that it contains the plasmid addiction genes phd and doc; pSSN6 contains clpXCc; and pSSN3 carries clpXEc. Growth of the following E. coli strains was monitored: W3110/pGB2ts/pSSN6 plus isopropyl-β-d-thiogalactopyranoside (IPTG) (▴; negative control); W3110/pGB2ts::phd-doc/pSSN6 plus IPTG (■; positive control); SSN1/pGB2ts/pSSN6 plus IPTG (▵); SSN1/pGB2ts::phd-doc/pSSN3 plus IPTG (□); SSN1/pGB2ts::phd-doc/pSSN6 plus IPTG (●); and SSN1/pGB2ts::phd-doc/pSSN6 (○). (B) Immunoblot analysis with an anti-ClpX serum and extracts of E. coli strains expressing ClpXCc and ClpXEc. Equal amounts of total protein from the following strains were analyzed: W3110/pMR20 (lane 1); W3110/pSSN3 (lane 2); W3110/pSSN6 (lane 3); SSN1/pMR20 (lane 4); SSN1/pSSN3 (lane 5); and SSN1/pSSN6 (lane 6). The band corresponding to ClpX is marked by an arrow.