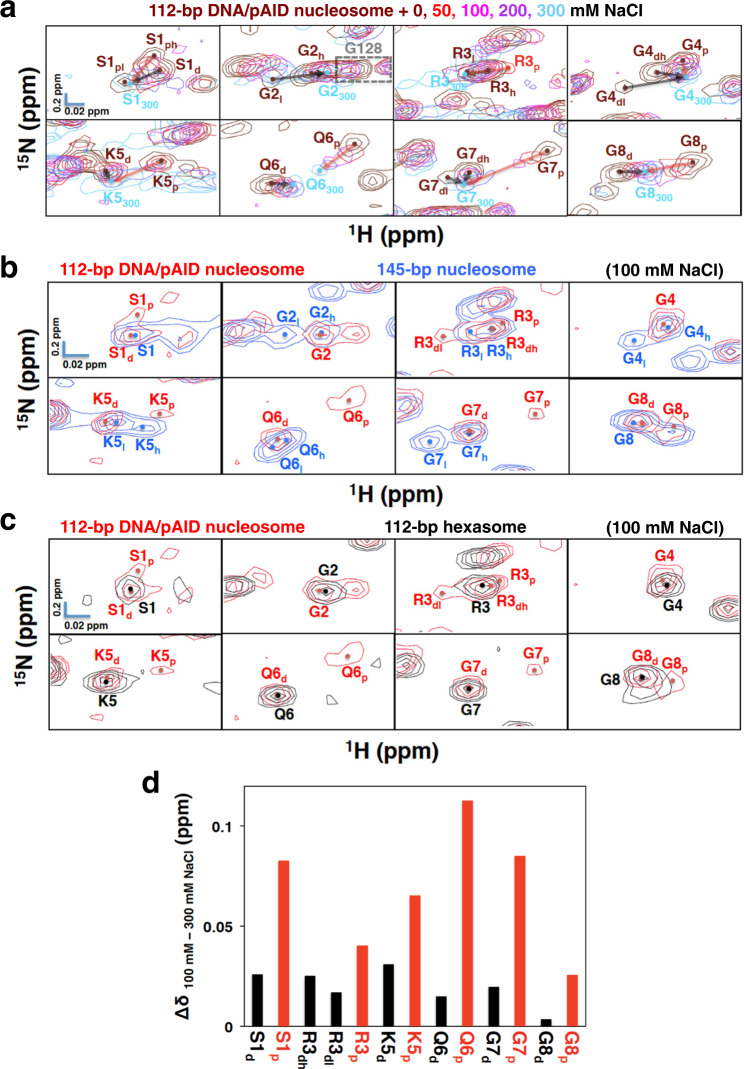
Fig. 4. Conformational comparison of the H2A N-tails under the physiological salt condition.
a Ionic strength dependence of signals of the H2A N-tail in the 112-bp DNA/pAID nucleosome upon NaCl titration at 0 M (brown), 50 mM (red), 100 mM (pink), 200 mM (purple), and 300 mM (cyan). NMR signals are divided into panels by amino acid residue. Residues are labeled in the color corresponding to the NaCl condition. Residues of the signal at 300 mM NaCl are designated by subscript 300. Red and black arrows indicate the chemical shift changes of the pAID and DNA side signals, respectively, between 0 or 50 mM and 300 mM NaCl. pAID and DNA side signals are designated by subscript p and d, respectively. High- and low-field components of the signals are designated by subscript h and l, respectively. b, c Expanded signal comparison between the 112-bp DNA/pAID nucleosome (red) and the 145-bp nucleosome (blue) (b) or the 112-bp hexasome (black) (c) at 100 mM NaCl. NMR signals are divided into panels by amino acid residue (Ser1–Gly8 of H2A). Signal assignments in the 112-bp DNA/pAID nucleosome, 145-bp nucleosome, and 112-bp hexasome are labeled in red, blue, and black, respectively. Filled circles indicate each signal center. d Histogram showing chemical shift differences between 100 mM and 300 mM NaCl of the pAID and DNA side signals in the H2A N-tail of the 112-bp DNA/ pAID nucleosome. Chemical shift differences are plotted for pAID side (red) and DNA side (black) residues in the H2A N-tail.