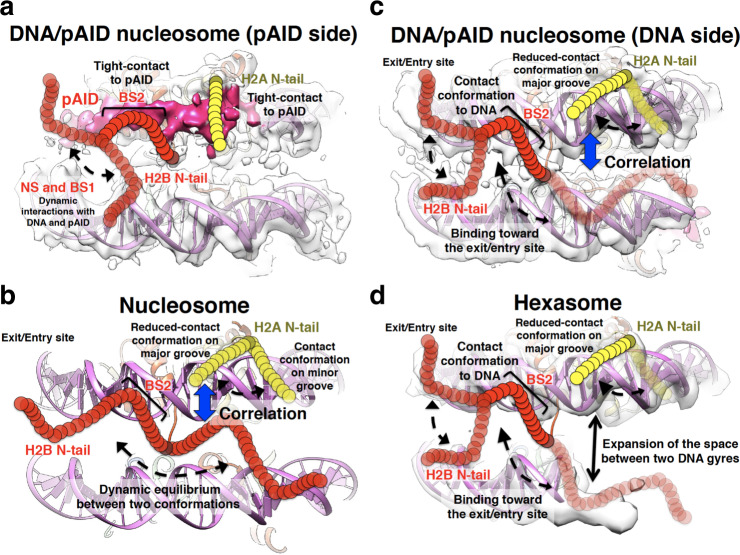
Fig. 8. Summary of the dynamic conformations of the H2A and H2B N-tails.
a–d Dynamic structures of the H2A and H2B N-tails in the DNA/pAID nucleosome on the pAID (a) and DNA (c) sides, in nucleosome (b), and in hexasome (d), colored as in Fig. 1. In b, the canonical nucleosome structure (PDB ID: 2CV5) is shown. In a, c, the EM density map of the 112-bp DNA/pAID nucleosome (EMD-9639) is superimposed on the nucleosome structure (PDB ID: 2CV5) lacking the 33-bp DNA. In d, the EM density map of the 112-bp hexasome (EMD-6939) is superimposed on the nucleosome structure (PDB ID: 2CV5) lacking the 43-bp DNA and one H2A/H2B dimer. Yellow and red circular chains denote the H2A and H2B N-tails, respectively. Dotted arrows represent the dynamic behavior of the H2A and H2B N-tails. Black arrow represents expansion of the space between two DNA gyres in hexasome, as shown in the cryo-EM structure of the 112-bp hexasome10. Blue arrows represent conformational correlation between the H2A N-tail and the H2B N-tail.