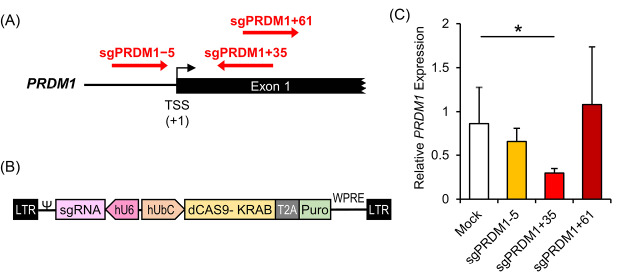
Fig. 3.
Knock-down of the transcription factor PR/SET domain 1 (PRDM1) gene by designed CRISPR interference (CRISPRi) constructs in human T cells. A) The sgRNA target sites near the PRDM1 transcription start site (TSS). The PRDM1 gene is located on chromosome 6. sgPRDM1-5 targets five nucleotides downstream of the TSS; sgPRDM1+35 targets 35 nucleotides upstream of the TSS; and sgPRDM1+61 targets 61 nucleotides upstream of the TSS. B) The CRISPRi construct to knock-down PRDM1. The vector is created to knock-down the PRDM1 gene by the designed sgRNA and the CAS9-KRAB enzyme. By pairing dCas9 with a sequence-specific sgRNA, the dCas9–sgRNA complex can interfere with transcription elongation by blocking RNA polymerase (Pol). It can also impede transcription initiation by disrupting transcription factor binding. In order to achieve enhanced repression, the Krüppel-associated box (KRAB) is fused to the carboxyl terminus of dCas9. Together with a target-specific sgRNA, the dCas9–KRAB fusion proteins can efficiently repress endogenous genes. The puromycin resistant gene expression product is segregated by T2A, a self-cleaving peptide, from the other products of the vector. C) RT-qPCR data depicts expression of the PRDM1 gene. The bar graph shows mRNA expression in different T cell groups transduced with CRISPRi constructs that comprise the designed sgRNAs. The three constructs perform differently in reducing the expression of PRDM1 mRNA. sgPRDM1+35 dramatically decreased PRDM1 gene expression at the mRNA level. The data are presented as mean ± SD, *P < 0.05, n = 3.