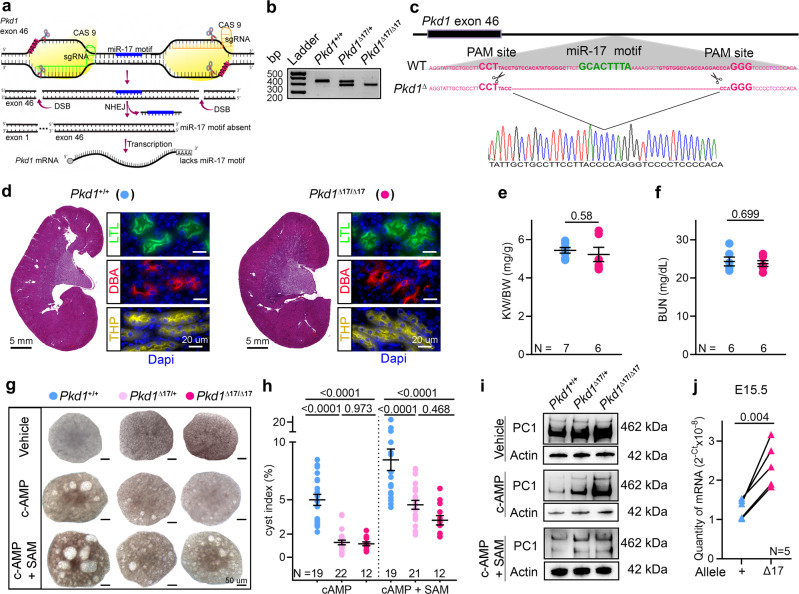
Fig. 1. Pkd1 mRNA is cis-repressed via its 3′-UTR miR-17 binding motif.
a Graphic illustration of the CRISPR/Cas9 approach used to delete the miR-17 motif from Pkd1 3′-UTR (Pkd1Δ17). b PCR products obtained after amplification of tail DNA from mice with indicated genotypes. The lower band represents the Δ17 deletion. n = 3 for all genotypes. c 3′-UTR nucleotide sequence of wildtype (WT) and Pkd1Δ17 alleles. The miR-17 binding motif and sgRNA PAM sites are highlighted in bold green and pink, respectively. The pink dashed line indicates the deleted nucleotides in Pkd1Δ17. Sanger sequencing chromatogram depicting the nucleotide sequence of the Pkd1Δ17 allele is shown. d H&E staining, Lotus Tetragonolobus Lectin labeling (LTL, a proximal tubule marker), Tamm-Horsfall protein immunostaining (THP, a loop of Henle maker), and Dolichos Biflorus Agglutinin labeling (DBA, a collecting duct marker) showing normal kidney histology in 8-week-old Pkd1+/+ and Pkd1Δ17/Δ17 mice. e–f Normal kidney-weight-to-body-weight (KW/BW) and serum blood urea nitrogen (BUN) levels in 8-week-old Pkd1+/+ and Pkd1Δ17/Δ17 mice. g–h Images and cyst index quantification of E13.5 Pkd1+/+, Pkd1Δ17/+, and Pkd1Δ17/Δ17 kidneys grown for four days in culture media containing vehicle, 100 uM cAMP, or 100 uM cAMP plus 250 uM SAM. i Immunoblots depicting PC1 expression in Pkd1+/+, Pkd1Δ17/+, and Pkd1Δ17/Δ17 ex-vivo kidneys treated with vehicle, cAMP, or cAMP plus SAM. Actin is used as the loading control. j Allele-specific qRT-PCR showing the quantity of Pkd1 mRNAs produced by the wildtype (+) and Δ17 alleles in E15.5 Pkd1Δ17/+ kidneys (n = 5). Error bars indicate SEM. Statistical analysis: Two-tailed Student’s t-test (e, f); One-way ANOVA, Tukey’s multiple-comparisons test (h); Two-tailed Paired t-test (j). Source data are provided as a Source Data file.