Figure 4.
The presence of m1Ψ in mRNA does not increase amino acid misincorporation frequency during translation in human cells
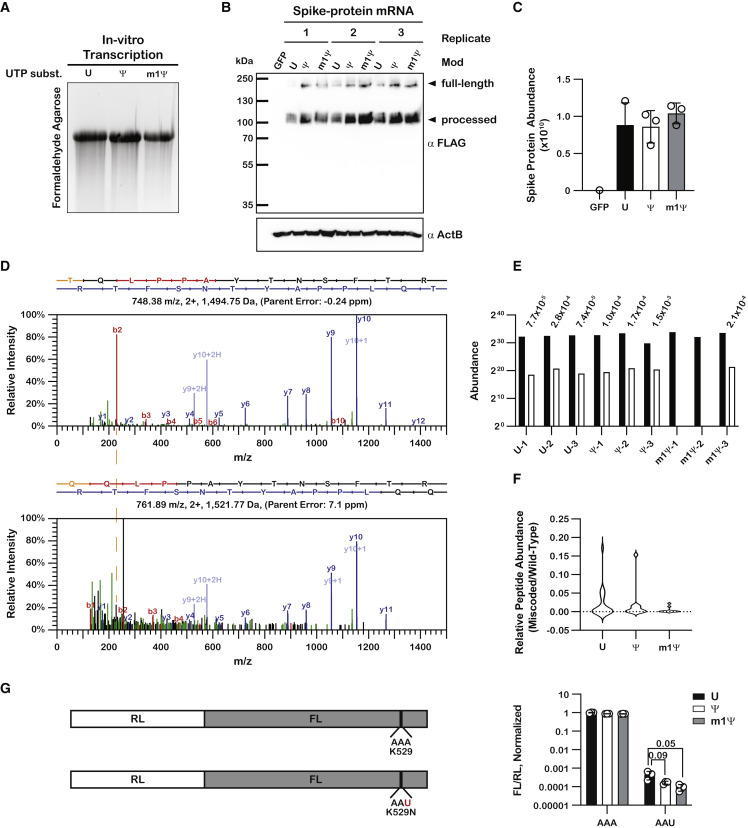
(A) UV-transillumination image of an ethidium bromide-stained formaldehyde agarose gel used to visualize the mRNA constructs transfected into HEK 293 cells.
(B) Immunoblot analysis of total cell lysate from transfected HEK 293 cells. The SARS-CoV-2 spike protein is known to undergo post-translational processing, and products corresponding to their respective sizes can be seen, as denoted by the arrows.
(C) Bar graphs showing the abundance of the wild-type SARS-CoV-2 spike protein isolated from on-bead digestion, as determined by label-free quantitation from LC-MS/MS analysis.
(D) Fragmentation spectra of the wild-type peptide TQLPPAYTNSFTR and its substituent miscoded product, QQLPPAYTNSFTR. b and y ions are denoted in red and blue, respectively, while the substituted amino acid is denoted in orange. The difference in the m/z for the b2–b4 peaks between the wild-type peptide (top) and miscoded product (bottom) corresponds to a threonine to glutamine substitution. The dashed orange line indicates the shift in the mass of the b2 ion. The nominal mass difference between threonine and glutamine is 27 Da.
(E) Bar graphs showing the abundance of TQLPPAYTNSFTR and its miscoded substituent, QQLPPAYTNSFTR. The miscoded peptide was not detected in samples m1Ψ-1 and m1Ψ-2. The relative peptide abundance is denoted above each pair.
(F) Violin plots showing the distribution of relative peptide abundances for miscoded spike protein peptides translated from U, Ψ-containing, or m1Ψ-containing mRNA. Only those peptides where both the wild-type and the miscoded species were detected were included.
(G) On the left is a schematic of the dual-luciferase reporter system used to assess miscoding frequency in HEK 293 cells. On the right are bar graphs showing normalized luminescence values for U, Ψ-containing, and m1Ψ-containing mRNA. Plotted are the average values of three biological replicates with error bars representing the standard deviation around the mean. Unpaired t tests did not show statistically significant differences in miscoding frequency between the U, Ψ-containing, and m1Ψ-containing mRNA.