Figure 7.
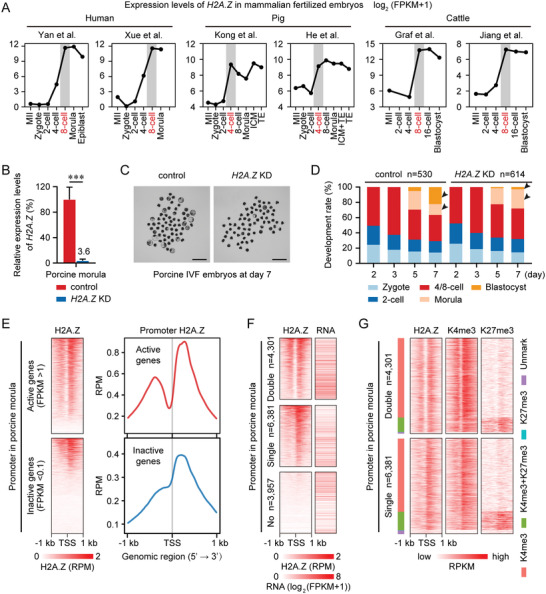
H2A.Z exhibits a conserved function in porcine early embryos. A) Line graphs showing the expression levels of H2A.Z in human, porcine and bovine MII oocytes and early embryos by RNA‐seq. RNA‐seq data for human,[ 15 , 32 ] pig[ 33 ] and cattle[ 34 ] are from previous publications. Developmental stage marked in red and gray shades indicates the corresponding ZGA stage. B) Bar charts showing the relative expression levels of H2A.Z in control and H2A.Z KD porcine embryos at the morula stage. These embryos are derived by IVF. H2A.Z expression levels in control morulae are set as 100%, and the relative H2A.Z expression levels in H2A.Z KD morulae are shown as a percentage. Error bars represent the SD in 4–5 replicates. ***P <0.001; two‐sided Student's t‐test. C) Representative images of control and H2A.Z KD porcine embryos at day 7 after fertilization. One representative image from four independent experiments is shown. Scale bar, 500 µm. D) Bar charts showing the development rate of control and H2A.Z KD porcine embryos at days 2, 3, 5, and 7 after fertilization. The numbers of embryos analyzed from four independent experiments are shown. Arrows point out the developmental differences between control and H2A.Z KD embryos. E) Heatmaps (left) and profiles (right) showing the H2A.Z enrichment at active (top) and inactive (bottom) gene promoters in porcine IVF embryos at the morula stage. Each row represents a promoter region (TSS ± 1 kb) and is ordered descending by H2A.Z enrichment. Two replicates of H2A.Z ULI‐NChIP‐seq are generated using porcine morulae. RNA‐seq data for porcine morulae are from an unpublished report (Genome Sequence Archive accession number: CRA004237). F) Heatmaps showing the H2A.Z enrichment and RNA levels at promoters with “Double”, “Single” and “No” H2A.Z peaks in porcine morulae. The numbers of genes with “Double”, “Single” and “No” H2A.Z peaks in porcine morulae are shown. Each row represents a promoter region and is ordered descending by H2A.Z enrichment. G) Heatmaps showing the enrichment of H2A.Z, H3K4me3, and H3K27me3 at promoters with “Double” and “Single” H2A.Z peaks in porcine morulae. Promoters are also divided into four clusters based on their histone modification patterns as follows: H3K4me3 (red), H3K27me3 (cyan), bivalent mark (H3K4me3+H3K27me3, green) and Unmark (purple). H3K4me3 and H3K27me3 ChIP‐seq data for porcine morulae are from an unpublished report (Genome Sequence Archive accession number: CRA003606). Two replicates of H3K4me3 and H3K27me3 ULI‐NChIP‐seq are generated using porcine morulae.
