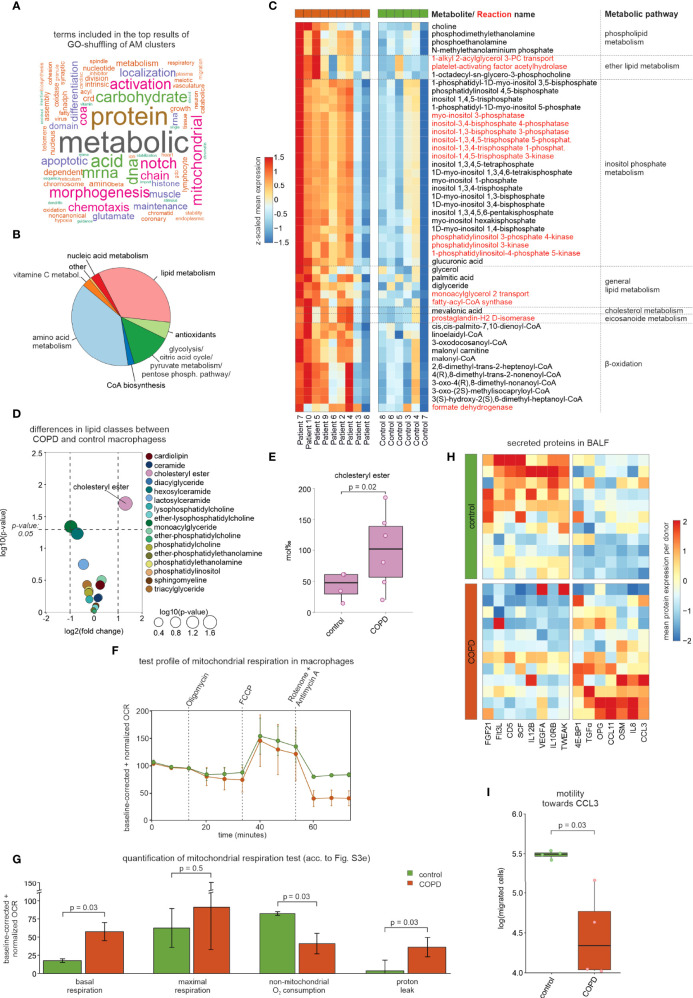
Figure 3.
Modeling of the metabolic landscape and alterations in macrophages. (A) Word cloud of the most common words in the top predicted terms of the GO-shuffling approach across all macrophage clusters. (B) Compass results of the modeled metabolic landscape in macrophages. The pie chart summarizes and categorizes the predicted metabolites and pathways that are significantly different between COPD and control. (C) Heatmap showing the metabolites and pathways that were predicted by Compass as altered in COPD and that were associated with lipid metabolism. Metabolites are shown in black and reactions in red. Columns and rows of the heat map are sorted by hierarchical clustering. (D) Volcano plot visualization of log2 fold changes and negative log10 p-values (Wilcoxon rank sum test) of lipid class levels between COPD and control macrophages obtained by lipidomics analysis. (E) Box plot with marked median values of cholesteryl ester proportions with the representation of individual donors. (F) Evaluation of mitochondrial function via the time-dependent course of the oxygen consumption rate (OCR) in macrophages using baseline-corrected values. Error bars indicate the standard deviations (control n = 2, COPD n = 3). Dashed arrows represent the injection of various compounds (shown at the top of the plot) used to assess different aspects of mitochondrial function (according to Figure S3 E). (G) Bar plots showing quantifications of different aspects of mitochondrial function inferred from the OCR measurement in Figure 3F (according to Figure S3 E; error bars indicating the standard deviation; statistics based on t-test). (H) Heatmap representation of proteins detected in BALF with a p-value < 0.1 according to the Wilcoxon rank sum test between COPD patients and control donors (control n = 11, COPD n = 12). The mean protein expression (identified by Olink Proteomics) per donor is represented as a z-transformed value (across all donors). Columns of the heatmap are sorted by hierarchical clustering. (I) Quantification of the migratory capability of macrophages towards CCL3 displayed in a box plot with marked median values and the representation of individual donors (control n = 4, COPD n = 4; error bars indicating the standard deviation; statistics based on t-test). BALF, bronchoalveolar lavage fluid; OCR, oxygen consumption rate.