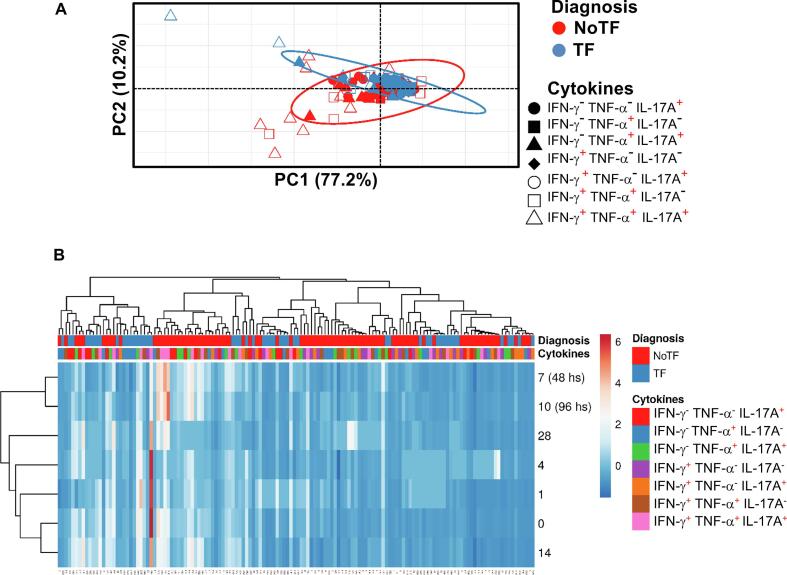
Fig. 5.
Hierarchical clustering of MAIT cell functions using principal component analysis (PCA). The three MAIT cell functions (IFN-γ, TNF-α, and IL-17A production) were analyzed using the FCOM deconvolution tool. FCOM data of the 7 possible combinations were used to perform an unsupervised PCA analysis. All data from NoTF and TF participants were merged for combined analysis, generating a matrix of the 7 possible phenotypes of cytokine producing MAIT cell subsets from the 20 participants at 7 different time points. (A) PCA. X and Y axis show principal component 1 and principal component 2, which account for 77.2% and 10.2% of the total variance, respectively. Prediction ellipses are such that with a probability of 0.95, a new observation from the same group will fall inside the ellipse (N = 140 data points). (B) Heatmap. Rows are centered; unit variance scaling was applied to rows. Imputation was used for missing value estimation. Both rows and columns are clustered using correlation distance and average linkage. 7 rows, 140 columns. PCA was generated using absolute MAIT cell numbers per microliter of peripheral blood.