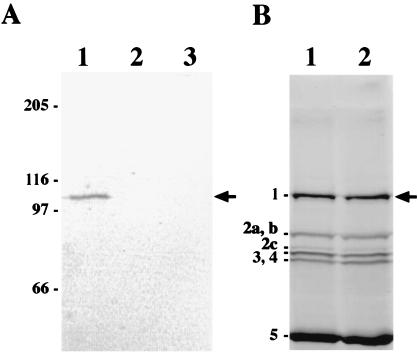
FIG. 1.
Immunodetection and penicillin-binding activity of PBP1-FLAG. (A) Western blot of PBP1-FLAG. Membranes from cells grown at 37°C in 100 ml of 2× YT medium were prepared and proteins were solubilized in SDS sample buffer and subjected to SDS–10% PAGE as described in Materials and Methods; about 40 μg of protein was loaded into each well of the gel. The gel was transferred to a polyvinylidene difluoride membrane, which was probed with 4 μg of monoclonal anti-FLAG antibody (Sigma) per ml, and bound antibodies were visualized by using alkaline phosphatase-conjugated secondary antibodies and a colorimetric alkaline phosphatase substrate as described in Materials and Methods. Lanes: 1, membrane proteins from strain LP27 expressing PBP1-FLAG; 2, cytoplasmic proteins from strain LP27; and 3, membrane proteins from wild-type cells (PS832). Molecular mass markers are shown in kilodaltons on the left. The arrow indicates the position of PBP1-FLAG. (B) Penicillin-binding activity of PBP1-FLAG. Membranes were prepared as described for panel A and incubated with FLU-C6-APA for 30 min at 30°C as described (46), proteins (60 μg per well) were separated by SDS–10% PAGE, and PBPs were visualized with a fluorimager. Lanes: 1, membrane proteins from strain LP27 expressing PBP1-FLAG; and 2, membrane proteins from wild-type cells (PS832). The positions of the major vegetative B. subtilis PBPs are shown on the left. The arrow indicates the position of PBP1 and PBP1-FLAG (due to the large size of PBP1, we could not observe any size difference between PBP1 and PBP1-FLAG).