Abstract
Targeting the programmed death 1/programmed death ligand 1 (PD-1/PD-L1) pathway with immunotherapy has revolutionized the treatment of many cancers. Somatic tumor mutational burden (TMB) and T-cell–inflamed gene expression profile (GEP) are clinically validated pan-tumor genomic biomarkers that can predict responsiveness to anti-PD-1/PD-L1 monotherapy in many tumor types. We analyzed the association between these biomarkers and the efficacy of PD-1 inhibitor in 11 commonly used preclinical syngeneic tumor mouse models using murinized rat anti-mouse PD-1 DX400 antibody muDX400, a surrogate for pembrolizumab. Response to muDX400 treatment was broadly classified into three categories: highly responsive, partially responsive, and intrinsically resistant to therapy. Molecular and cellular profiling validated differences in immune cell infiltration and activation in the tumor microenvironment of muDX400-responsive tumors. Baseline and on-treatment genomic analysis showed an association between TMB, murine T-cell–inflamed gene expression profile (murine-GEP), and response to muDX400 treatment. We extended our analysis to investigate a canonical set of cancer and immune biology-related gene signatures, including signatures of angiogenesis, myeloid-derived suppressor cells, and stromal/epithelial-to-mesenchymal transition/TGFβ biology previously shown to be inversely associated with the clinical efficacy of immune checkpoint blockade. Finally, we evaluated the association between murine-GEP and preclinical efficacy with standard-of-care chemotherapy or antiangiogenic agents that previously demonstrated promising clinical activity, in combination with muDX400. Our profiling studies begin to elucidate the underlying biological mechanisms of response and resistance to PD-1/PD-L1 blockade represented by these models, thereby providing insight into which models are most appropriate for the evaluation of orthogonal combination strategies.
Introduction
Immunotherapy has changed the way patients with cancer are being treated, providing durable tumor regression and overall survival (OS; refs. 1, 2). Despite the success of immune checkpoint blockade, only a subset of patients with certain tumor types will achieve a durable response with programmed death 1/programmed death ligand 1 (PD-1/PD-L1) inhibitors (3–5). Combination therapy approaches may be key to improving clinical outcomes for these patients. Furthermore, understanding the underlying immunosuppressive mechanisms within the tumor microenvironment (TME) that differentiate response from lack of response to PD-1/PD-L1 inhibitors, including immune infiltration and stromal profiles, is necessary for the development of novel interventions and strategies that enhance antitumor immune activity and durability of response.
Clinical studies have yielded insight into predictive biomarkers of response to PD-1/PD-L1 inhibitors. Although PD-L1 expression is a broadly used biomarker of response and a companion diagnostic for PD-1/PD-L1 inhibitors across multiple tumor types (6–8), it is only one component of a much more complex network within the TME. A defined IFNγ-related, 18-gene T-cell–inflamed gene expression profile (T-cellinfGEP) indicative of an immunogenic TME is associated with favorable clinical response to the PD-1 inhibitor pembrolizumab across multiple tumor types (9). Additionally, high tumor mutational burden (TMB) correlates with benefit from PD-1/PD-L1 inhibition (10–12). Stratification of biomarker-defined subgroups based on the joint utility of TMB and T-cellinfGEP has identified discrete patterns of responsiveness to pembrolizumab, collectively uncovering underlying targetable biology associated with response and resistance to pembrolizumab monotherapy (13). Further, several exploratory gene signatures are associated with resistance to pembrolizumab monotherapy, including angiogenesis, myeloid-derived suppressor cell (MDSC), and stromal/epithelial-to-mesenchymal transition (EMT)/TGFβ biology-related signatures (14, 15). These ongoing translational efforts inform the discovery of new targets and orthogonal combination strategies.
Preclinical models complement clinical activities to elucidate which patients respond to PD-1/PD-L1 inhibitors by enabling understanding of the underlying biology and antitumor activity. Syngeneic tumor models are an experimental tool of choice to study treatment-related immune modulation of the TME and have yielded insight into the development of immunotherapies. However, there are known limitations in the translation from mouse to human, including immunologic differences (16–18); how these differences affect our ability to model clinically relevant mechanistic biomarkers of response to PD-1/PD-L1 inhibitors has not been fully explored. Herein, we elucidate which biological mechanisms can or cannot be appropriately and productively tested in syngeneic tumor models. Using the rodent-surrogate pembrolizumab antibody muDX400, we evaluated the relationship between TMB and murine-surrogate T-cell–inflamed gene expression profile (murine-GEP) and response. We also evaluated gene signatures in pretreatment biopsies from mice subsequently treated with muDX400 and standard-of-care agents that have shown promising combination activity in clinical settings.
Methods
Cancer cell lines and culture
Mouse tumor cell lines 4T1, CT26, RENCA, CM3, B16-F10, EMT6, LL/2, and TC1 were obtained from ATCC; MC38 from the NCI DTP; MBT2 from RIKEN; and MB49 from the O’Donnell laboratory at the University of Iowa (Iowa City, IA). All cell lines were maintained as monolayers in DMEM or RPMI-1640 supplemented with 10% heat-inactivated FBS and grown at 37°C and 5% CO2. All tumor cells were routinely subcultured two to three times per week depending on the growth rate and split ratio. All cell lines were authenticated using short tandem repeat profiling and were confirmed to be free of Mycoplasma and mouse pathogens (e.g., C. bovis) by PCR testing between 2018 and 2021.
Syngeneic tumor models
Seven- to 9-week-old female mice were purchased (C57BL/6, DBA/2, and C3H from the Jackson Laboratory; BALB/c from Taconic Biosciences). All procedures involving the care and use of animals were reviewed and approved by the Institutional Animal Care and Use Committee of Merck & Co., Inc., Kenilworth, NJ, USA and were conducted in accordance with the regulations and guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care. Mice were anesthetized and inoculated subcutaneously into the right lower flank with a single-cell suspension of ≥95% viable cells in 0.1 mL of serum-free culture media. Tumors and body weights were measured twice weekly using a caliper and the formula for tumor volume [V = 0.5 (a × b2); a and b are the long and short diameters of the tumor, respectively].
Treatments
Rat anti-mouse PD-1 antibody (rDX400) antibody was murinized (muDX400) for in vivo use (18). Mouse antibodies were dosed intraperitoneally (i.p.) at 5 mg/kg every 5 days for ≤5 doses. Commercially available anti-PD-1 reagent clones RMP1-14 [rat immunoglobulin G (IgG) 2a] and J43 (hamster IgG) were used for comparison with muDX400. Monotherapy or combination treatments began when subcutaneous tumors reached an average of 100 to 200 mm3 within 8 to 12 days, depending on the model. Mice were pair-matched into treatment groups based on initial tumor size, and researchers were unblinded to treatments. For chemotherapy combination studies, carboplatin was given i.p. at 40 mg/kg every 7 days, and pemetrexed was given i.p. at 200 mg/kg every 7 days (doses predetermined by MTD). muIgG1 or muDX400 were given i.p. at 10 mg/kg every 5 days. Lenvatinib was dosed by oral gavage at 10 mg/kg every day. All efficacy studies were conducted with n = 10 mice per treatment group to provide statistical power and account for model variability. The data presented represent at least three separate studies.
Study outcomes
Median OS and molecular and cellular characterization of tumor models, including TMB and a 15-gene murine GEP, were assessed. Tumor volume was calculated using the formula 0.5 × length × width2, where length was the longer dimension. Tumor growth inhibition (TGI) was calculated using the formula [(Ct–C0)–(Tt–T0)]/(Ct–C0) × 100, where Ct represents the mean tumor volume of the control group at time (t), C0 the mean tumor volume of the control group at t0, Tt the mean tumor volume of the treatment group at t, and T0 the mean tumor volume of the treatment group at t0.
Flow cytometry
Freshly dissected tumors were minced and dissociated in digestion media (gentleMACS system; Miltenyi Biotec) using the 37_m_TDK program. Cell suspensions were washed and counted, and 2 × 106 cells were incubated with fixable viability stain 510 dye (BD Biosciences) or eFluor 506 (eBioscience), followed by blocking with anti-CD16/CD32b antibody for 10 minutes on ice. Cells were then stained with fluorochrome-conjugated surface antibodies prepared in a 1× staining buffer (BioLegend) and brilliant stain buffer (BD Biosciences) solution for 30 minutes on ice. For detection of intracellular expression of Foxp3, cells were permeabilized and fixed with Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer's instructions and incubated with Foxp3 antibody for 30 minutes. Stained samples were acquired on a Fortessa cytometer (BD Biosciences) with BD FACSDiva software and analyzed using FCS Express (De Novo Software) or Flow Jo (BD Biosciences). See Supplementary Methods for additional details.
IHC
IHC was performed on formalin-fixed paraffin-embedded tissue sections with the following antibodies: anti-mouse CD3, clone CD3-12 (AbD Serotec); anti-mouse PD-L1, clone MEB077.6H4.181 (Merck & Co., Inc., Kenilworth, NJ, USA); and anti-mouse PD-1, goat polyclonal (R&D Systems). See Supplementary Methods for additional details.
RNA extraction, targeted RNA profiling, RNA sequencing, and WES
For targeted RNA profiling and RNA sequencing, tumor-bearing mice were established and treated similarly to those in efficacy studies with samples collected at specified timepoints. Tumors (between 50 and 600 mm3, dependent on model) were excised and snap-frozen in liquid nitrogen and stored at −80°C until RNA isolation. See Supplementary Methods for additional details (GEO accession No. GSE168846).
Murine-GEP signature score was generated by computing the z-score [of log2 fragments per kilobase million (FPKM) values] for each of the 15 genes across the subset of samples (baseline or following muIgG1 isotype control or muDX400 treatment, as indicated), multiplying the z-score for Cd276 by –1 and then taking the mean, as described previously (13). The three human leukocyte antigen genes in the human T-cellinfGEP signature (13) were excluded because of cross-species differences. A similar method was used to compute the scores for the additional gene signatures (15). For evaluation of the additional gene signatures (15) in the mouse data set, the heat map compares the pairwise Pearson correlation coefficients evaluated over baseline tumor samples. Gene expression in samples was based on log2(0.01+FPKM) for protein-coding genes. For evaluation of reference signatures in the human data set, the heat map compares pairwise Spearman correlation coefficients evaluated over solid tumor samples after adjusting for tumor type. Gene expression in samples was based on log10(0.01+FPKM) for protein-coding genes after global normalization by the within-sample 75th percentile. Comparison of the pairwise correlations between mice and humans was performed in MATLAB R2019a (MathWorks). Heat maps and box plots were created using R (v3.5.2).
For WES, genomic DNA (gDNA) was extracted from cell lines grown in vitro using standard protocols. gDNA was sequenced using the SureSelect Mouse Exon Kit (Agilent) with 90 base paired-end reads on the HiSeq 2500 (Illumina). See Supplementary Methods for additional details.
Results
Differential response to muDX400 treatment across syngeneic tumor models
Anti-mouse PD-1 mAbs were generated to enable pharmacologic studies and mechanistic assessment of PD-1 inhibition in mouse models. The monoclonal rat anti-mouse PD-1 antibody (rDX400) and murinized version (muDX400) were validated as surrogates for pembrolizumab. The biophysical characteristics of muDX400 and pembrolizumab have been previously compared (18). In the commonly used MC38 colon adenocarcinoma tumor model, administration of the parental rDX400 antibody resulted in more durable regression and prolonged survival compared with the commonly used anti-PD-1 clones RMP1-14 and J43 (Supplementary Fig. S1A and S1B). To diminish the generation of anti-rat antibodies in the mouse models, muDX400 was used for additional efficacy and profiling efforts. In 11 widely used syngeneic tumor models, muDX400 monotherapy elicited a range of model-specific responses with differential sensitivity to PD-1 inhibition independent of tumor origin and mouse strain (Fig. 1). Models were broadly categorized as highly responsive (≥70% TGI with multiple complete regressions), partially responsive (30%–70% TGI with occasional complete regressions), and intrinsically resistant (<30% TGI) to muDX400 monotherapy. These classifications were based on standardized starting tumor volumes per model across >10 studies. MC38, MBT2, and CM3 tumor models showed multiple partial and complete regressions, RENCA, CT26, MB49, and EMT6 tumor models were partially responsive, and 4T1, TC1, B16-F10, and LL/2 tumor models exhibited intrinsic resistance to muDX400 treatment (Fig. 1A). Assessment of PD-1 and PD-L1 protein expression by IHC from baseline (100 mm3) tumor samples showed variable expression levels, with enrichment of PD-1/PD-L1-low-expressing tumors in the muDX400-resistant category (Supplementary Table S1 shows the protein and RNA expression levels of PD-1 and PD-L1).
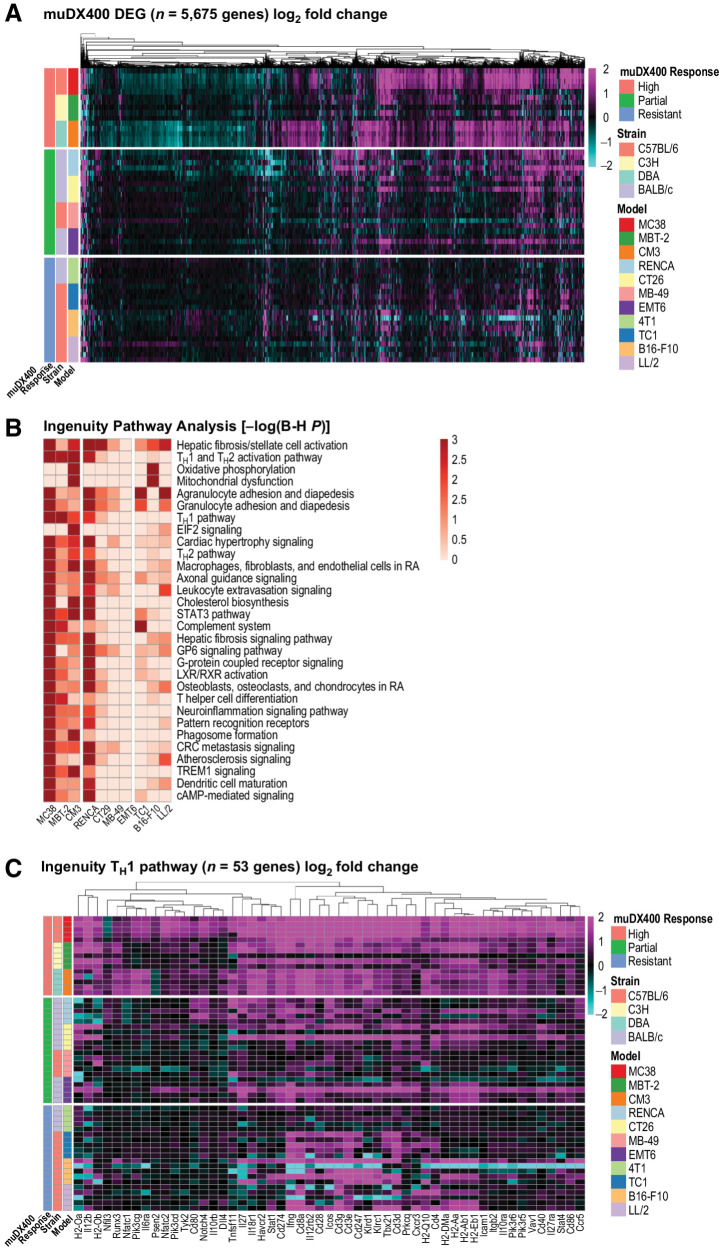
Figure 1.
Differential responsiveness to anti-PD-1 therapy (muDX400) in syngeneic tumor models. A, Spider plots depicting tumor growth across syngeneic tumor models classified as highly responsive (>70% TGI with multiple CRs), partially responsive (30%–70% TGI with occasional CRs), or resistant (<30% TGI) to murinized rat anti-mouse PD-1 DX400 antibody muDX400. CR (absence of detectable tumor) and PR (>30% tumor shrinkage) are specified. Single-agent treatment with 5 mg/kg muDX400 or 5 mg/kg muIgG1 isotype control was administered when subcutaneous tumors reached 100 mm3 on average, denoted as day 0. Black arrows indicate specific dates on which doses were administered intraperitoneally, with dosing every 5 days for up to 5 total doses. Data shown are representative of at least 10 independent experiments with n ≥ 10 mice per group. B, Summary of muDX400 response categorization including tumor origin and mouse strain for each syngeneic tumor model. CR, complete response; muIgG1, mouse immunoglobulin G1; PD-1, programmed death 1; TGI, tumor growth inhibition.
Immune activation following muDX400 treatment is predominantly restricted to the tumor
We evaluated molecular correlates of response to muDX400 in the MC38 tumor model. Targeted gene expression analysis (RT-qPCR), using a panel of 437 immune gene markers, identified an altered molecular signature with significant upregulation of genes involved in monocyte, B-cell, and T-cell activation and differentiation at day 8 (4 days after a second dose of muDX400) in the tumor tissue but not peripheral blood or draining lymph nodes (dLNs) of responding mice (Supplementary Fig. S1C and S1D). These results align with receptor occupancy studies demonstrating that antitumor response to muDX400 in the MC38 tumor model is driven by the level of PD-1 blockade in the tumor but not blood or dLNs (19). Efficient PD-1 receptor occupancy on tumor-infiltrating T cells is important for antitumor activity of PD-1 therapy (18). We observed more robust immune gene changes in the tumor after multiple doses of muDX400 (Supplementary Fig. S1D). To determine whether these observations were model specific, we performed bulk RNA-sequencing analysis on tumors from muDX400 highly responsive (MC38), partially responsive (CT26), and resistant (TC1) models. Similar to the RT-qPCR analysis on MC38 tumors, more pronounced gene changes occurred in the tumor tissue on day 8 than on day 4 after treatment across all three models (Supplementary Fig. S1E). The observed dynamic alteration of the immune landscape correlated with response to muDX400 (Supplementary Fig. S1E). Because this preliminary experiment showed a change in gene expression most apparent after two doses of muDX400, we chose day 8 for the analysis of subsequent tissue collections.
Transcriptional upregulation of genes associated with IFNγ signaling and TH1 cells in muDX400-responsive models
Next, we expanded the investigation of transcriptome-wide changes in tumors with bulk RNA sequencing across all 11 syngeneic tumor models (Fig. 2). On day 8 after muDX400 treatment, the highest number of differentially expressed genes in the tumor tissue was observed in models most responsive to treatment (MC38, MBT2, CM3) compared with partially responsive (RENCA, CT26, MB49) and resistant models (4T1, TC1, B16-F10, LL/2). Key immune gene expression changes included upregulation of genes related to IFNγ signaling and TH1 differentiation (Ifng, Stat1, Il27ra, Stat4, Tbx21, Il12rb2, Cxcr3, Ccl5), T-cell activation, and cytolytic activity (Il2ra, Cd3d, Cd3e, Cd3g, Cd8a, Ccr7, Icos, Zap70, Lck, Lat, Cd28, Gzmb, Prf1), antigen presentation and dendritic cell maturation (Cd80, Cd86, Cd40, Cd83, Il12a, Il12b), chemokine signaling and leukocyte extravasation (Ccl3, Ccl4, Ccl5, Ccl6, Cxcl9, Cxcl10, Cxcr4, Cxcr6, Itgal, Itgb2), and adaptive immune resistance (Pdcd1, Lag3, Cd274, Pdcd1lg2, Ido1, Tigit; Fig. 2; Supplementary Table S2). We further observed that several genes associated with a progenitor-exhausted CD8+ T-cell state were upregulated after muDX400 treatment in responsive but not resistant models, including Tcf7, Slamf6, Ccr7, and Tox (Supplementary Table S2). The effects of gene upregulation corresponded to the extent of TGI and, despite some variability, were generally consistent across models in the same muDX400 response category (Figs. 1A and 2A). Mice in muDX400 partially responsive models demonstrated variable gene expression changes (Fig. 2A), similar to the variability in muDX400 efficacy response (Fig. 1A). Ingenuity Pathway Analysis across tumor models highlighted several immune-related pathways enriched by muDX400 at day 8, including the TH1 pathway, dendritic cell maturation, leukocyte extravasation signaling, and TH1 differentiation (Fig. 2B). Effects of muDX400 on gene regulation within the Ingenuity TH1 pathway are shown in Fig. 2C. Highly responsive models showed the most profound effects, including upregulation of genes such as Stat1, Cxcr3, Il12rb2, and Ifng; a few mice in the partially responsive models also exhibited gene expression regulation similar to that of the highly responsive models. Collectively, these results demonstrate that muDX400 recapitulates the activation of T-cell cytolytic activity observed in the clinical setting with PD-1 blockade.
Figure 2.
GEP of syngeneic tumor models following treatment with anti-PD-1 (muDX400). A, RNA sequencing of bulk tumors excised from syngeneic tumor models following single-agent treatment with 5 mg/kg muDX400 or muIgG1 isotype control antibody, 4 days after the second dose. Shown in the heat map are the 5,675 genes differentially regulated by muDX400 compared with isotype control antibody in any of the 11 tumor models (1.5× and FDR <0.05, base mean >20; see Supplementary Table S3). The color gradient represents the log2-fold change of each mouse (each row) treated with muDX400 compared with the corresponding pooled control mice treated with muIgG1 isotype control antibody at baseline (±log2-fold). Sample details (such as barcodes and replicate numbers) are shown in Supplementary Table S1. B, Ingenuity Pathway Analysis of the 5,675 genes (comparison analysis) identified the canonical pathways most significantly regulated by muDX400 treatment across the syngeneic tumor models. Shown in the heat map are the top 30 pathways. muDX400 did not result in significantly enriched pathways in the 4T1 model, which was therefore excluded from the heat map. The color gradient represents pathway enrichment P values. C, Shown in the heat map are the genes in the TH1 Ingenuity canonical pathway (log2-fold change of muDX400 treatment compared with isotype control). The color gradient is as shown in A. B-H P, Benjamini–Hochberg P value; DEG, differentially expressed genes; FDR, false discovery rate; GEP, gene expression profile; muIgG1, mouse immunoglobulin G1; PD-1, programmed death 1; TH1, type 1 helper T cell.
Cellular phenotypic analysis shows differences in tumor immune cell infiltrate in muDX400-responsive versus -resistant models
To compare changes in RNA and protein expression, immune cell infiltration was analyzed using multiparameter flow cytometry. In baseline tumor samples (100 mm3) across the 11 models, the highest frequencies of tumor-infiltrating CD45+ immune cells, including CD4+ and CD8+ T cells, were observed in highly responsive models, whereas MDSCs were enriched in tumors resistant to muDX400 monotherapy (Supplementary Fig. S2). On-treatment tumor samples from highly responsive models exhibited significant increases in CD8+ T-cell frequency, determined as a proportion of CD45+ cells (Supplementary Fig. S2C–S2E); this increase was characterized by a higher CD8 to regulatory T-cell ratio (Supplementary Fig. S2F). These results align with previous findings that a T-cell-infiltrated TME is an important feature of PD-1 inhibitor-responsive tumors. Frequencies of other immune cell populations remained largely unchanged after muDX400 treatment in partially responsive and resistant models (Supplementary Fig. S2C). Similar results were observed by IHC, with increased infiltration of CD3+ T cells at baseline and following treatment with muDX400 in highly responsive MC38 tumors, but not treatment-resistant B16-F10 tumors (Supplementary Fig. S2G). These results show alignment between molecular and cellular tumor immune changes observed with muDX400 treatment and validate muDX400 as a useful tool in the evaluation of immune-mediated effects of PD-1 blockade.
Association between murine-GEP and TMB and response to muDX400 treatment at baseline and posttreatment
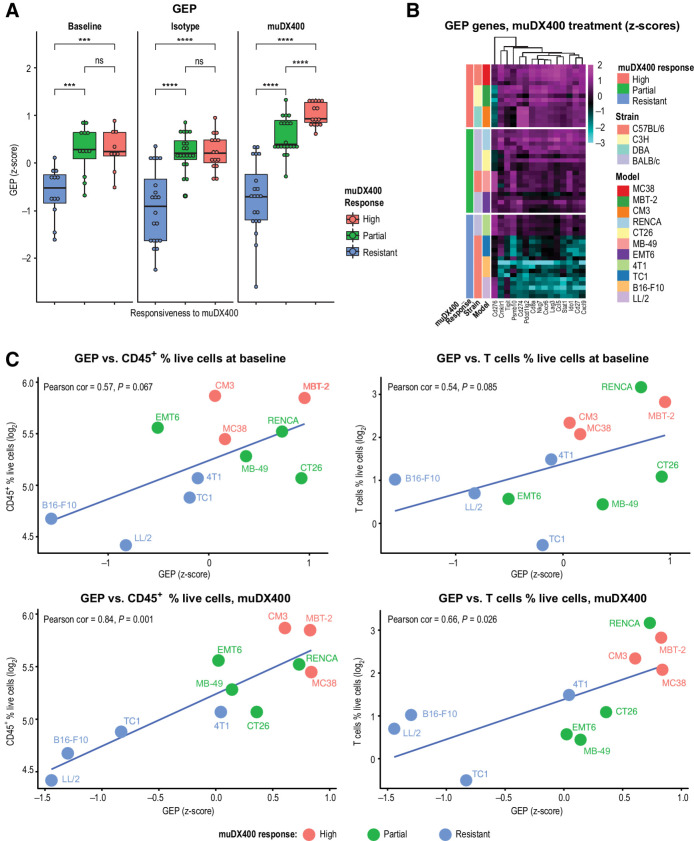
We evaluated concordance between clinically derived biomarkers predictive of sensitivity and resistance to anti-PD-1 therapy across multiple tumor types, with efficacy based on our muDX400 response categorization (Fig. 1). This included the T-cellinfGEP composed of genes involved in T-cell activation signals, antigen-presenting cell machinery, and IFNγ signaling pathways prospectively validated in melanoma and predictive of response or resistance to pembrolizumab monotherapy in multiple tumor types (20). To elucidate the relationship between T-cellinfGEP and muDX400 response categorization, we adopted a surrogate murine-GEP signature corresponding to the T-cellinfGEP signature. Murine-GEP score was associated with responsiveness to muDX400 monotherapy in syngeneic tumor models (Fig. 3). At baseline (100-mm3 tumors), murine-GEP score was significantly higher in muDX400-responsive compared with muDX400-resistant models (Fig. 3A). Analysis of day 8 on-treatment samples revealed a significantly higher murine-GEP score in responsive models, with an overall increase in murine-GEP score compared with isotype-treated controls (Fig. 3A and B). Expression of the 15 murine-GEP genes across the muDX400 response categories is shown in Supplementary Fig. S3. Moreover, murine-GEP score showed a positive trend at baseline, or a significant positive correlation after muDX400 treatment with the percentage of tumor-infiltrating CD45+ cells or CD3+ T cells at baseline (Fig. 3C; CD45+ cells: Pearson correlation = 0.84, P = 0.0013; T cells: Pearson correlation = 0.66, P = 0.026). A similar positive trend was observed for baseline murine-GEP score and frequencies of tumor-infiltrating CD4+/CD8+ T cells and CD11b+ myeloid cells, whereas MDSCs exhibited a trend toward a negative association (Supplementary Fig. S2). We further assessed the relationship between murine-GEP and clinically relevant biomarkers of response to PD-1 inhibition, including PD-1, PD-L1, and PD-L2 (Fig. 4A; Supplementary Fig. S3). muDX400-responsive models were characterized by higher Pdcd1 (Pd-1), Cd274 (Pd-l1), and Cd273 (Pd-l2) mRNA expression levels at baseline and following treatment with muDX400. In addition, TMB and neoantigens were higher in muDX400-responsive models (Fig. 4B). Collectively, these reverse-translational studies reveal that clinically derived biomarkers associated with response to anti-PD-1 inhibition are also associated with muDX400 response in syngeneic tumor mouse models.
Figure 3.
Immune-related murine-GEP signature associates with responsiveness to anti-PD-1 (muDX400) in syngeneic tumor models. RNA-sequencing analysis was performed on five independent tumors across 11 syngeneic tumor models after anti-PD-1 treatment. Tumors were harvested 4 days following two doses of muDX400 or muIgG1 control antibody (treatment administered on day 0 and day 4). Models were classified as resistant, partial, or high as previously defined. A, A 15-gene murine-GEP score, corresponding to the T-cellinfGEP signature (9), without the three HLA genes, was evaluated before and after muDX400 or after muIgG1 isotype control antibody treatment represented as mean z-score (of log2 FPKM values). Significance was determined by t test (NS, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). B, Heat map of the genes from the 15-gene murine-GEP score. The color gradient shows the z-score (of log2 FPKM values) of each gene (columns) across the mouse samples (rows) treated with muDX400. C, Correlation of murine-GEP and immune cell frequencies at baseline. The 15-gene murine-GEP scores (mean z-scores of the log2 FPKM values) at baseline or after muDX400 treatment are plotted against the percentage of CD45+ cells or T cells (of total live cells, at baseline) in each tumor model. FPKM, fragments per kilobase million; GEP, gene expression profile; HLA, human leukocyte antigen; muIgG1, mouse immunoglobulin G1; ns, not significant; PD-1, programmed death 1; T-cellinfGEP, T-cell-inflamed gene expression profile.
Figure 4.

Evaluation of clinically validated biomarkers in syngeneic tumor models. RNA-sequencing analysis was performed on five independent tumors across 11 syngeneic tumor mouse models after anti-PD-1 treatment. Tumors were harvested 4 days following the second dose of muDX400 or muIgG1 control antibody (treatment administered on day 0 and day 4). Models were classified as resistant, partial, or high as previously defined. A, Boxplots of Cd274 (Pdl1), depicting the absolute gene expression (log2 FPKM) across individual mouse in each tumor model grouped by responsiveness to muDX400 and by treatment. Significance determined by t test (NS, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). B, TMB (all somatic nonsynonymous mutations) and predicted neoantigens (total and expressed at the transcriptional level) are shown across the 11 mouse tumor cell lines (grown in vitro). None of the comparisons reached statistical significance (by t test) but they did show a trend (NS, P > 0.05). C, Individual count data. FPKM, fragments per kilobase million; muIgG1, mouse immunoglobulin G1; NS, not sufficient; PD-1, programmed death 1; TMB, tumor mutational burden.
Assessment of exploratory gene signatures associated with anti-PD-1 resistance
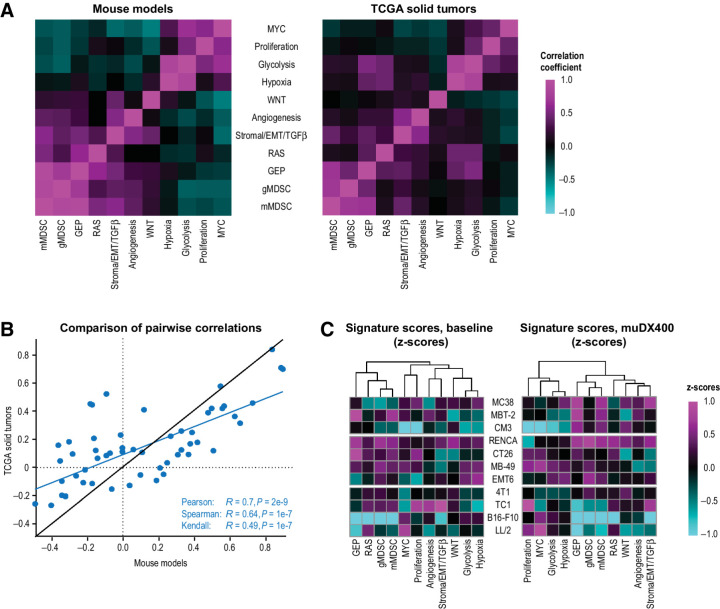
To better understand the translational utility of these models, we examined a set of canonical gene signatures, including immune- and cancer-related signatures of angiogenesis, MDSC, and stromal/EMT/TGFβ-related biology associated with pembrolizumab resistance (15). We generated exploratory murine gene signatures derived from mouse orthologous genes and compared their expression in mouse tumors with those in solid tumors from patients with cancer using the Cancer Genome Atlas database. A heat map shows the comparison of correlation coefficients between the signatures for each species in Fig. 5A; a scatterplot shows the correlation of those coefficients in Fig. 5B. This comparison revealed an overall similarity in the global expression patterns of these signatures between the two species, demonstrating conserved features of human tumor resistance biology at the level of syngeneic tumor models. T-cellinfGEP at baseline in human tumors has demonstrated a robust positive association with OS on pembrolizumab monotherapy (13); in this study, we observed a positive association between murine-GEP and muDX400 response (Fig. 5C). Whereas signatures for monocytic MDSC (mMDSC), angiogenesis, and stroma/EMT/TGFβ biology showed evidence of negative association with response to pembrolizumab in human tumors at baseline (15), the high variability in the expression of these signatures in mouse syngeneic tumors did not reveal any association with response to muDX400 monotherapy (Fig. 5C). However, resistance signatures for gMDSC-, mMDSC-, and hypoxia-related tumor biology exhibited higher expression at baseline and following muDX400 treatment in partially responsive models relative to resistant and highly responsive models (Fig. 5C), suggesting that a hypoxic TME enriched in MDSCs may subvert the activity of muDX400 in partially responsive tumors. Collectively, these results provide a framework for the rational selection of syngeneic tumor models in the evaluation of discrete biological mechanisms underlying resistance to PD-1/PD-L1 inhibition.
Figure 5.
Exploratory GEPs associated with resistance to anti-PD-1 inhibition (muDX400) show similar expression patterns in syngeneic tumor models. A, Additional reference signatures (15) are summarized in the covariance plots for mouse tumors (present study) and human solid tumors (TCGA). Heat maps compare the pairwise correlation coefficients for all the signatures, evaluated across tumor samples. The color gradient represents the pairwise correlations within each species. The correlation between mouse and human, of the pairwise correlation coefficients within each species (from A), is shown in the scatterplot in B. Expression of the reference signatures, at baseline or after muDX400 treatment, is shown in the heat maps across the tumor models in C. The color gradient represents the z-scores (of log2 FPKM values) of each signature (columns) across the mouse models (rows). EMT, epithelial-mesenchymal transition; FPKM, fragments per kilobase million; GEP, gene expression profile; gMDSC, granulocytic myeloid–derived suppressor cells; mMDSC, monocytic myeloid–derived suppressor cells; TCGA, The Cancer Genome Atlas; TGF-β, transforming growth factor β.
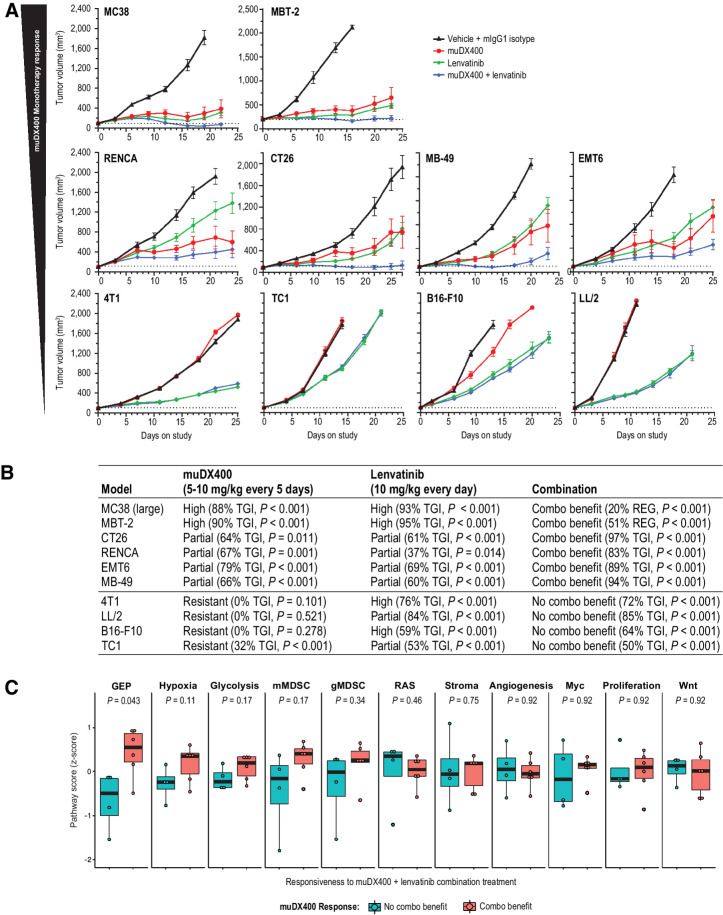
Evaluation of preclinical efficacy with standard-of-care and antiangiogenic combinations with muDX400 and association with murine-GEP
Clinically, anti-PD-1 plus antiangiogenic therapy or platinum-containing chemotherapy is emerging as a promising combination treatment characterized by robust antitumor activity across multiple tumor types (21–23). To understand whether preclinical signatures at baseline provide any predictive utility for combination benefit, we compared combination therapy of muDX400 plus lenvatinib with monotherapy across 10 syngeneic tumor models (Fig. 6A). Although all models were responsive to lenvatinib monotherapy, only highly or partially muDX400-responsive models exhibited benefit from combination therapy whereby efficacy was statistically improved over monotherapy with either agent (Fig. 6B). Furthermore, elevated baseline murine-GEP score (Wilcox P = 0.043) was associated with models that exhibited benefit from lenvatinib and anti-PD-1 combination therapy (Fig. 6C). Although resistance signature scores (e.g., hypoxia, glycolysis, mMDSC) trended toward being higher in models in which lenvatinib and anti-PD-1 combination therapy showed enhanced antitumor activity, none passed adjustment for multiple hypothesis testing. Taken together, these findings suggest that the murine-GEP score is the most reliable predictor of whether efficacy of anti-PD-1 plus lenvatinib is greater than monotherapy with either agent in these models. We also evaluated the combination of muDX400 and platinum chemotherapy across a subset of syngeneic mouse models with differential muDX400 response profiles (Supplementary Fig. S4A). Unlike combination therapy with an antiangiogenic agent, the combination of chemotherapy and PD-1 blockade showed benefit in muDX400-responsive [MC38 (300 mm2), RENCA, EMT6] and muDX400-resistant (4T1) models (Supplementary Fig. S4A). In contrast, we did not observe a significant benefit with combination therapy in the TC1 or CT26 models (Supplementary Fig. S4A). Chemotherapy combination efficacy and murine-GEP scores for these studies are summarized in Supplementary Fig. S4B. Although additional syngeneic models are needed to better understand the relationship between murine-GEP and response to platinum-based chemotherapy regimens, these studies suggest that murine-GEP may be unnecessary for response to combination therapies with orthogonal mechanisms.
Figure 6.
Antitumor efficacy of lenvatinib plus anti-PD-1 (muDX400) in syngeneic tumor models. A, Tumor growth curves for 10 subcutaneous syngeneic models treated with 5 mg/kg IgG1 isotype control antibody Q5D, 5 mg/kg muDX400 Q5D, 10 mg/kg lenvatinib QD, or muDX400 plus lenvatinib. Dotted lines represent tumor size at initiation of treatment. TGI and observations of complete or partial tumor regressions are indicated for each respective tumor regimen. B, The 10 models in A were classified as displaying improved efficacy when both agents were administered (combination benefit) or as no different from monotherapy with either agent (no combination benefit). The monotherapy response classification was as described in Fig. 1. C, Reference signature scores (Fig. 5) were evaluated at baseline and represented as mean z-score (of log2 FPKM values). P values shown represent the nominal Wilcox test P values. None of the signatures passed adjustment for multiple hypothesis testing. CR, complete response; Combo benefit, combination benefit; FPKM, fragments per kilobase million; GEP, gene expression profile; gMDSC, granulocytic myeloid–derived suppressor cells; muIgG1, mouse immunoglobulin G1; mMDSC, monocytic myeloid–derived suppressor cells; PD-1, programmed death 1; PR, partial response; QD, every day; Q5D, every 5 days; TGI, tumor growth inhibition.
Data availability
Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA (MSD) is committed to providing qualified scientific researchers access to anonymized data and clinical study reports from the company's clinical trials for the purpose of conducting legitimate scientific research. MSD is also obligated to protect the rights and privacy of trial participants and, as such, has a procedure in place for evaluating and fulfilling requests for sharing company clinical trial data with qualified external scientific researchers. The MSD data sharing website (http://engagezone.msd.com/ds_documentation.php) outlines the process and requirements for submitting a data request. Applications will be promptly assessed for completeness and policy compliance. Feasible requests will be reviewed by a committee of MSD subject matter experts to assess the scientific validity of the request and the qualifications of the requestors. In line with data privacy legislation, submitters of approved requests must enter into a standard data-sharing agreement with MSD before data access is granted. Data will be made available for request after product approval in the United States and EU or after product development is discontinued. There are circumstances that may prevent MSD from sharing requested data, including country or region-specific regulations. If the request is declined, it will be communicated to the investigator. Access to genetic or exploratory biomarker data requires a detailed, hypothesis-driven statistical analysis plan that is collaboratively developed by the requestor and MSD subject matter experts; after approval of the statistical analysis plan and execution of a data-sharing agreement, MSD will either perform the proposed analyses and share the results with the requestor or will construct biomarker covariates and add them to a file with clinical data that is uploaded to an analysis portal so that the requestor can perform the proposed analyses.
Discussion
Preclinical tumor mouse models play an important role in the development of anticancer therapeutics and are used to determine dose selection, drug treatment efficacy, mechanisms of action or biology, and combination strategies. Understanding the translatability of these models and their predictive power and limitations is key to designing rational anticancer therapies. Immune-competent syngeneic tumor models, which are genetically distinct and exhibit a range of immunogenicity, enable the interrogation of innate and adaptive immunity in the TME (24–28). However, these models have their limitations, and their translational utility has been a subject of ongoing investigations (24–28). Despite these advances, it remains unclear whether functional gene signatures defined in the clinical setting can be prospectively validated in syngeneic tumor models. The reverse-translational studies described here show that several features of pretreatment tumor biopsies predictive of clinical outcome to anti-PD-1 therapy in patients with cancer were similarly associated with the degree of response to muDX400 in syngeneic tumor models. We show an association between response to muDX400 monotherapy and murine-GEP in pretreatment tumor biopsies and report findings on molecular modulation of the TME after anti-PD-1 treatment. A higher murine-GEP score was observed after treatment in muDX400-sensitive models, consistent with clinical observations and the limited ability of PD-1/PD-L1 inhibition to turn “cold” tumors “hot.” Pretreatment tumor biopsies showed that muDX400-sensitive models exhibited higher expression levels of several clinical biomarkers associated with response to anti-PD-1 therapy (e.g., PD-L1, TMB) and increased infiltration of CD8+ T lymphocytes compared with resistant models (Figs. 3 and 4; Supplementary Fig. S2). These observations mirror correlates of response to pembrolizumab in T-cellinfGEP/TMB biomarker-defined patient subgroups, in which T-cellinfGEP/TMBhigh tumors are enriched for PD-1 inhibition responders (13). By validating the association between murine-GEP and response to muDX400 therapy, our study positions syngeneic tumor models in a clinically biomarker-defined framework that can enable rational design and evaluation of immunotherapies and combination strategies.
To better understand the applicability of syngeneic tumor models in the evaluation of mechanisms of resistance to PD-1/PD-L1 therapy, we generated an exploratory set of murine gene signatures associated with clinical resistance to pembrolizumab (15). There was a high degree of similarity in global expression patterns of these signatures between murine and human solid tumors but substantial diversity within the 11 syngeneic tumor models at baseline and after treatment with muDX400 (Fig. 5). These findings are important for several reasons. First, although the number of mouse tumors in our study was limited, pairwise correlations demonstrate a consistent level of resistance signature expression in tumors between species, uncovering an additional layer of resistance biology that is appropriately modeled at the level of syngeneic tumors. Second, the diversity of resistance mechanisms and intermodel heterogeneity provides a conceptual framework to select models that may be applicable in the evaluation of specific mechanisms of resistance to PD-1/PD-L1 inhibition. Finally, by examining the modulation of these resistance signatures in muDX400 posttreatment tumor biopsies, we can begin to rationalize combinations that may prevent resistance in an individually syngeneic tumor model-tailored and resistance signature-dependent manner. For example, the mMDSC signature negatively impacts anti-PD-1 efficacy in the clinic but does not appear to be expressed at higher levels in muDX400-resistant models at baseline, or after treatment with muDX400, compared with responsive models (Fig. 5C). In contrast, mMDSC and gMDSC signature scores were higher in muDX400 partially responsive models at baseline; this pattern of expression was maintained or increased in posttreatment tumor biopsies, suggesting that antitumor immunity may be blunted by expanding MDSCs in these models. Our observations of higher mMDSC or gMDSC resistance signature expression in the partially responsive RENCA model is directly supported by a recent study demonstrating that as RENCA tumors increase in size, immune infiltrate generally decreases but MDSC density dramatically increases in the growing RENCA tumor (24). As these tumors grow, excess production of VEGF contributes to vasculature remodeling and immunosuppression (24), which is also consistent with a higher angiogenesis resistance signature expression following muDX400 treatment (Fig. 5C). Although these signatures may provide important information about mechanisms of inherent resistance to anti-PD-1 therapy in syngeneic tumor models, it remains unclear whether inherent and acquired resistance mechanisms share similar underlying biology. Moreover, whether mechanisms of acquired resistance can be appropriately modeled by syngeneic tumor models remains to be determined.
Recent studies have demonstrated that a population of progenitor-exhausted CD8+ T cells expressing the transcription factor Tcf1 preferentially respond to anti-PD-1 monotherapy in syngeneic tumor models (29–31). Elevated frequencies of progenitor-exhausted CD8+ T cells in melanoma tumor lesions of human subjects were predictive of response and survival to anti-PD-1 monotherapy (29, 32). Interestingly, we observed upregulation of several genes associated with progenitor-exhausted CD8+ T-cell identity in muDX400 responsive models, suggesting that pretreatment tumor biopsies of muDX400-responsive models may be enriched in these cells. Although single-cell RNA-sequencing studies will be required to definitively establish the presence of this population in muDX400-sensitive tumors, expansion of CD8+ T cells was the major compositional change in tumor models highly responsive to muDX400 monotherapy supports this notion (Supplementary Fig. S4). It remains unknown whether enrichment of progenitor-exhausted CD8+ T cells is necessary and sufficient for enhanced antitumor activity observed in models highly responsive to muDX400.
Immunotherapy combined with nonimmunotherapy has shown marked improvement in response rates in multiple tumor types and in OS in non–small cell lung cancer (22, 23). Recent studies assessing the clinical activity of PD-1/PD-L1 inhibitor combination therapies indicate that a wide range of mechanisms of action show activity as measured by tumor response (22). It has been suggested that for most of these agents, combination activity can be attributed to the independent activity of each agent versus mathematically greater activity consistent with clinical synergy (23). Although preclinical data recommended these strategies, the trials were empirically based, combining drugs with known efficacy and minimal overlapping toxicity. For translational oncology, these results demand a reexamination of biomarkers such as PD-L1, TMB, and TcellinfGEP and demand reverse translational studies to further understand the mechanisms that help decipher nonresponse and resistance. Evaluation of patient samples is key for these efforts, but the ability to model mechanisms or biology of response and resistance in preclinical animal models could accelerate insights by allowing us to test hypotheses within a fixed immune context, potentially leading to the identification of new targets.
To understand the translatability of preclinical animal models, we evaluated the association between clinical signatures and efficacy using two of the most commonly tested combinations with PD-1/PD-L1 inhibitors [platinum chemotherapies and VEGF(R) agents] in syngeneic tumor models (22, 23). Lenvatinib, a multiple kinase inhibitor that inhibits the three main VEGFRs (1–3) and other proangiogenic and oncogenic pathway-related receptor tyrosine kinases, including FGFR1–4, platelet-derived growth factor receptor-α, KIT, and RET (33), has shown promising antitumor activity in combination with pembrolizumab in several solid tumor types and is emerging as a potentially novel regimen for tumors with limited therapeutic options (34, 35). In our study, lenvatinib therapy enhanced anti-PD-1 antitumor activity in syngeneic tumor models with higher baseline murine-GEP scores, suggesting that an underlying immune component may be associated with combination benefit (Fig. 6A). Previous molecular epidemiology analysis of gene signatures associated with vascular biology suggested that genes related to vascular biology was enriched in TMBlow/TcellinfGEPhigh tumors (13). It will be of interest to determine the utility of these biomarkers in lenvatinib plus PD-1/PD-L1 inhibitor combination therapy in future clinical and translational studies. Whether TcellinfGEP or TMB can identify patients who derive greater benefit from upfront combination approaches over anti-PD-1 monotherapy remains unknown. Evaluation of gene signatures representing key biological axes of expression beyond TcellinfGEP showed no obvious association with response to lenvatinib plus anti-PD-1 in the syngeneic tumor models tested (Fig. 6C). Evaluating how lenvatinib affects murine-GEP levels after anti-PD-1 treatment and determining whether that mechanism of action involves modulation of resistance signature expression will be important in unraveling mechanisms of lenvatinib combination benefit. Unlike the observed trend with VEGFR plus PD-1 inhibition, murine-GEP was not associated with response to platinum-based chemotherapy because both muDX400-responsive and muDX400-resistant models showed combination benefit (Supplementary Fig. S5). These observations are based on a limited number of syngeneic tumor models but suggest that murine-GEP may not be uniformly applicable as a predictive biomarker of response to all combinations, especially those involving orthogonal mechanisms.
In summary, our study provides a platform for the rational evaluation of anti-PD-1 immunotherapies and combination regimens through the validation of clinical biomarkers and gene signatures associated with response or resistance in syngeneic tumor models. A critical aspect of this work is its ability to position these models in a PD-1 resistance signature framework that provides a rationale for what mechanisms can be appropriately modeled across a diverse set of syngeneic tumor models. Although additional validation will be necessary to identify drivers of resistance in specific models, similarities in the global expression patterns of resistance signatures between human and mouse solid tumors support the translational use of these models in identifying combination approaches aimed at overcoming PD-1 resistance. Understanding the limitations of syngeneic tumor models will be equally important for the development of new models or optimal approaches that expand beyond syngeneic tumor models for the evaluation of future oncology drug strategies.
Supplementary Material
Acknowledgments
Medical writing and/or editorial assistance was provided by Holly C. Cappelli, PhD, CMPP, of ApotheCom. This assistance was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. We thank Alexander Kulyk and Jennifer Kawai Cho for assistance with the next-generation sequencing. This research was funded by Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
P. Georgiev reports personal fees from Merck & Co., Inc., Kenilworth, NJ, USA and employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA outside the submitted work. E.S. Muise reports personal fees from Merck & Co., Inc., Kenilworth, NJ, USA and employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA outside the submitted work. D.C. Wilson reports personal fees from Merck & Co., Inc., Kenilworth, NJ, USA and is a stockholder of Merck & Co., Inc., Kenilworth, NJ, USA during the conduct of the study. S. Sukumar reports employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and is a stockholder of Merck & Co., Inc., Kenilworth, NJ, USA during the conduct of the study. L. Chen reports employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA outside the submitted work. J.H. Yearley reports a patent 0241115 issued and a patent 10647771 issued. V. Sriram reports a patent 9,827,309 issued. M. Nebozhyn reports personal fees, non-financial support, and employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA during the conduct of the study and outside the submitted work. H.A. Hirsch reports that at the time of data generation, employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA; at the time of submission, employment at CRISPR Therapeutics; and at the time of acceptance, employment at IMV Inc. D.Y. Chiang reports personal fees from Novartis and personal fees from Bayer outside the submitted work. R. Cristescu reports employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA and is a stockholder of Merck & Co., Inc., Kenilworth, NJ, USA outside the submitted work; in addition, R. Cristescu has a patent for Angiogenesis and mMDSC Gene Expression Based Biomarker of Tumor Response to PD-1 Antagonists pending and a patent for WO 2020/167619 pending. B.J. Long is an employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA. T.K. McClanahan reports employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA outside the submitted work. E.M. Pinheiro reports employment from Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
P. Georgiev: Conceptualization, formal analysis, investigation, writing–original draft, writing–review and editing, interpretation of the results. E.S. Muise: Formal analysis, writing–original draft, writing–review and editing, interpretation of the results. D.E. Linn: Conceptualization, formal analysis, investigation, writing–original draft, writing–review and editing, interpretation of the results. M.C. Hinton: Formal analysis, investigation, writing–original draft, writing–review and editing. Y. Wang: Formal analysis, investigation, writing–original draft, interpretation of the results. M. Cai: Conceptualization, formal analysis, investigation, writing–review and editing. L. Cadzow: Conceptualization, investigation, writing–review and editing. D.C. Wilson: Formal analysis, writing–review and editing, interpretation of the results. S. Sukumar: Conceptualization, formal analysis, investigation, writing–review and editing, interpretation of the results. M. Caniga: Investigation, writing–review and editing. L. Chen: Formal analysis, writing–review and editing. H. Xiao: Formal analysis, writing–review and editing. J.H. Yearley: Formal analysis, investigation, writing–review and editing, interpretation of the results. V. Sriram: Conceptualization, writing–review and editing, interpretation of the results. M. Nebozhyn: Formal analysis, writing–review and editing. M. Sathe: Formal analysis, writing–review and editing, acquisition of the data, interpretation of the results. W.M. Blumenschein: Investigation, writing–original draft, interpretation of the results. K.S. Kerr: Investigation, writing–review and editing. H.A. Hirsch: Conceptualization, formal analysis, investigation. S. Javaid: Formal analysis, interpretation of the results. A.K. Olow: Formal analysis, writing–review and editing. L.Y. Moy: Formal analysis, writing–review and editing, interpretation of the results. D.Y. Chiang: Writing–review and editing, interpretation of the results. A. Loboda: Formal analysis, writing–review and editing, interpretation of the results. R. Cristescu: Writing–review and editing, interpretation of the results. S. Sadekova: Conceptualization, writing–review and editing. B.J. Long: Conceptualization, formal analysis, writing–original draft. T.K. McClanahan: Conceptualization, writing–original draft, writing–review and editing, interpretation of the results. E.M. Pinheiro: Conceptualization, formal analysis, writing–original draft, writing–review and editing, interpretation of the results.
References
- 1. Sharma P, Allison JP. The future of immune checkpoint therapy. Science 2015;348:56–61. [DOI] [PubMed] [Google Scholar]
- 2. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science 2018;359:1350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kalbasi A, Tariveranmoshabad M, Hakimi K, Kremer S, Campbell KM, Funes JM, et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci Transl Med 2020;12:eabb0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nowicki TS, Hu-Lieskovan S, Ribas A. Mechanisms of resistance to PD-1 and PD-L1 blockade. Cancer J 2018;24:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res 2014;20:5064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 2017;357:409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cristescu R, Nebozhyn M, Zhang C, Albright A, Kobie J, Huang L, et al. Transcriptomic determinants of response to pembrolizumab monotherapy across solid tumor types. Clin Cancer Res 2021;clincanres.3329.2021. doi: 10.1158/1078-0432.CCR-21-3329. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 16. Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol 2004;172:2731–8. [DOI] [PubMed] [Google Scholar]
- 17. Rongvaux A, Takizawa H, Strowig T, Willinger T, Eynon EE, Flavell RA, et al. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu Rev Immunol 2013;31:635–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature 2016;532:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Plitnick LM, Hutchins B, Dubey S, Li N, Amin RP, Born S, et al. A T-cell-dependent antibody response study using a murine surrogate anti-PD-1 monoclonal antibody as an alternative to a non-human primate model. J Immunotoxicol 2020;17:175–85. [DOI] [PubMed] [Google Scholar]
- 20. Ayers M, Nebozhyn M, Cristescu R, McClanahan TK, Perini R, Rubin E, et al. Molecular profiling of cohorts of tumor samples to guide clinical development of pembrolizumab as monotherapy. Clin Cancer Res 2019;25:1564–73. [DOI] [PubMed] [Google Scholar]
- 21. Taylor MH, Lee CH, Makker V, Rasco D, Dutcus CE, Wu J, et al. Phase IB/II trial of lenvatinib plus pembrolizumab in patients with advanced renal cell carcinoma, endometrial cancer, and other selected advanced solid tumors. J Clin Oncol 2020;38:1154–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schmidt EV. Developing combination strategies using PD-1 checkpoint inhibitors to treat cancer. Semin Immunopathol 2019;41:21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmidt EV, Chisamore MJ, Chaney MF, Maradeo ME, Anderson J, Baltus GA, et al. Assessment of clinical activity of PD-1 checkpoint inhibitor combination therapies reported in clinical trials. JAMA Network Open 2020;3:e1920833. [DOI] [PubMed] [Google Scholar]
- 24. Yu JW, Bhattacharya S, Yanamandra N, Kilian D, Shi H, Yadavilli S, et al. Tumor-immune profiling of murine syngeneic tumor models as a framework to guide mechanistic studies and predict therapy response in distinct tumor microenvironments. PLoS One 2018;13:e0206223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhong W, Myers JS, Wang F, Wang K, Lucas J, Rosfjord E, et al. Comparison of the molecular and cellular phenotypes of common mouse syngeneic models with human tumors. BMC Genomics 2020;21:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mosely SI, Prime JE, Sainson RC, Koopmann JO, Wang DY, Greenawalt DM, et al. Rational selection of syngeneic preclinical tumor models for immunotherapeutic drug discovery. Cancer Immunol Res 2017;5:29–41. [DOI] [PubMed] [Google Scholar]
- 27. Lechner MG, Karimi SS, Barry-Holson K, Angell TE, Murphy KA, Church CH, et al. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J Immunother 2013;36:477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen IX, Newcomer K, Pauken KE, Juneja VR, Naxerova K, Wu MW, et al. A bilateral tumor model identifies transcriptional programs associated with patient response to immune checkpoint blockade. Proc Natl Acad Sci U S A 2020;117:23684–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller AM, Lemke-Miltner C, Blackwell S, Tomanek-Chalkley A, Coleman K, Weiner G, et al. CMP-001, a virus-like particle containing immunostimulatory CpG-A, for treatment of peritoneal carcinomatosis of gastrointestinal and pancreatic cancers. Cancer Res 2019;79:3273. [Google Scholar]
- 30. Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 31. Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016;537:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamamoto Y, Matsui J, Matsushima T, Obaishi H, Miyazaki K, Nakamura K, et al. Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vasc Cell 2014;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tahara M, Schlumberger M, Elisei R, Habra MA, Kiyota N, Paschke R, et al. Exploratory analysis of biomarkers associated with clinical outcomes from the study of lenvatinib in differentiated cancer of the thyroid. Eur J Cancer 2017;75:213–21. [DOI] [PubMed] [Google Scholar]
- 35. Taylor MH, Schmidt EV, Dutcus C, Pinheiro EM, Funahashi Y, Lubiniecki G, et al. The LEAP program: lenvatinib plus pembrolizumab for the treatment of advanced solid tumors. Future Oncol 2021;17:637–48. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA (MSD) is committed to providing qualified scientific researchers access to anonymized data and clinical study reports from the company's clinical trials for the purpose of conducting legitimate scientific research. MSD is also obligated to protect the rights and privacy of trial participants and, as such, has a procedure in place for evaluating and fulfilling requests for sharing company clinical trial data with qualified external scientific researchers. The MSD data sharing website (http://engagezone.msd.com/ds_documentation.php) outlines the process and requirements for submitting a data request. Applications will be promptly assessed for completeness and policy compliance. Feasible requests will be reviewed by a committee of MSD subject matter experts to assess the scientific validity of the request and the qualifications of the requestors. In line with data privacy legislation, submitters of approved requests must enter into a standard data-sharing agreement with MSD before data access is granted. Data will be made available for request after product approval in the United States and EU or after product development is discontinued. There are circumstances that may prevent MSD from sharing requested data, including country or region-specific regulations. If the request is declined, it will be communicated to the investigator. Access to genetic or exploratory biomarker data requires a detailed, hypothesis-driven statistical analysis plan that is collaboratively developed by the requestor and MSD subject matter experts; after approval of the statistical analysis plan and execution of a data-sharing agreement, MSD will either perform the proposed analyses and share the results with the requestor or will construct biomarker covariates and add them to a file with clinical data that is uploaded to an analysis portal so that the requestor can perform the proposed analyses.