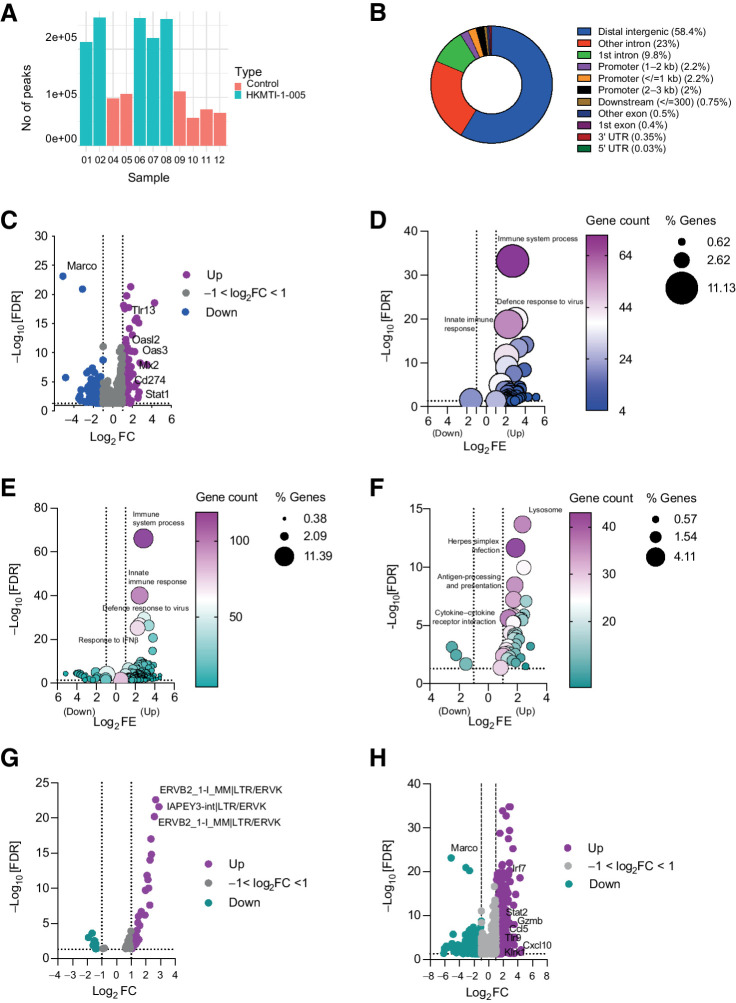
Figure 3.
ATAC-seq and RNA-seq on murine omental tumor deposits. A, Number of ATAC-seq peaks for control (n = 6) and HKMTI-1–005 samples (n = 5), before applying filtration criteria. One HKMTI-1–005 sample did not yield enough sequencing reads and was removed from analysis. B, Distribution of ATAC-seq peaks across the genome. C, Overlap of genes with ATAC-seq peaks showing increased chromatin accessibility that were also differentially expressed (n = 1,106) by RNA-seq following HKMTI-1–005 treatment in vivo (HKMTI-1–005 n = 7, control n = 7). FC, fold change. Purple color: Log2FC ≥ 1, gray color: −1 < Log2FC ≤ 1, and blue color: Log2FC ≤ −1. D, Biological processes (BP) sub-ontology for 1,053 genes from C that overlapped with gene expression signatures from DAVID online Functional Annotation Tool. Gene count denotes the number of genes found to overlap with genes within the respective signature and the dot size represents the percentage of these genes within the signature. FE, fold enrichment. E and F, DEG following HKTMI-1–005 classified by BP and KEGG sub-ontologies, respectively. Gene count denotes the number of genes found to overlap with genes within the respective signature and the dot size represents the percentage of these genes within the signature. G, Volcano plot showing differentially expressed ERVs, following HKMTI treatment (n = 7) versus control (n = 7). Purple color: Log2FC ≥ 1, gray color: −1 < Log2FC ≤ 1 and green color: Log2FC ≤ −1. H, Volcano plot showing individual DEG following HKMTI treatment (n = 7) versus control (n = 7) by RNA sequencing with an FDR <0.05. Purple color: Log2FC ≥ 1, gray color: −1 < Log2FC ≤ 1 and green color: Log2FC ≤ −1. Cellular response to IFNβ (GO:0035458, FDR = 7.65e−15) and response to virus (GO:0009615, FDR = 4.32e−14).