Figure 2.
Summary of key clinical parameters, treatment regimen, and sequencing results in patient 1
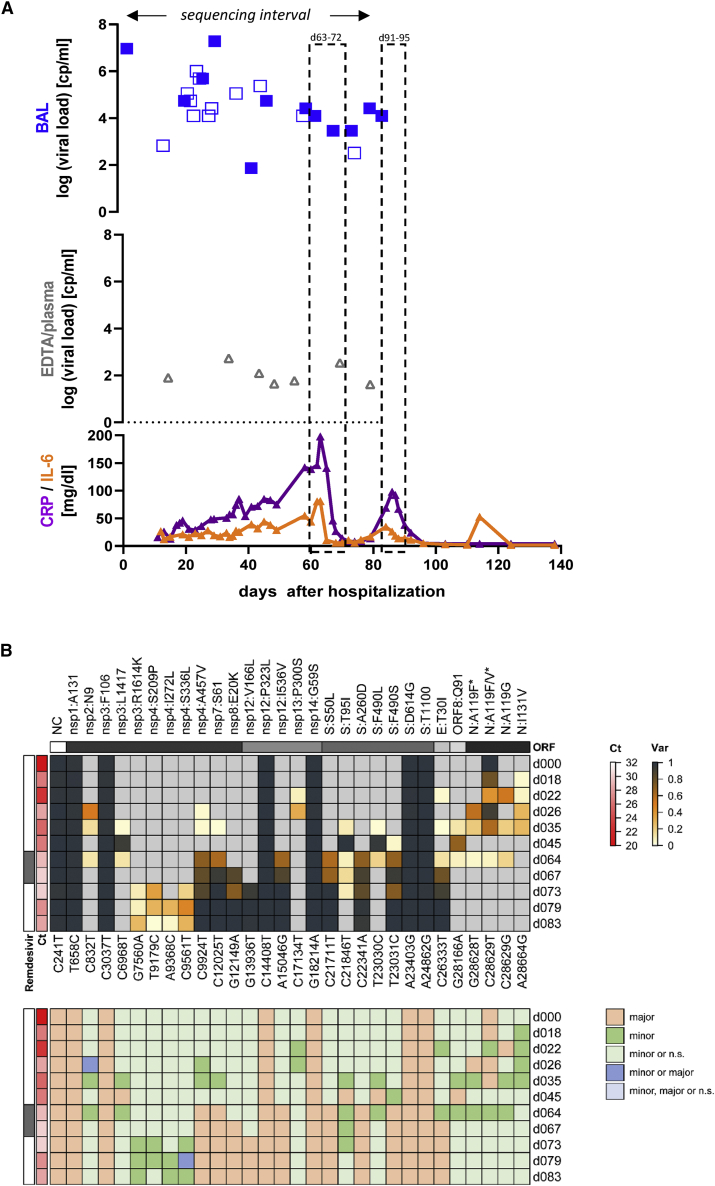
(A) Temporal representation of the infection interval indicating dates when respiratory samples were collected and whether they tested positive for SARS-CoV-2, along with remdesivir treatment dates (light gray boxes) and inflammatory markers (CRP and IL-6). CRP values are shown in green and IL-6 in turquoise. Viral loads are shown for respiratory samples (green diamonds) and blood samples (gray triangles). Filled symbols depict samples subjected for sequencing.
(B) Upper panel: Heatmap depicting the frequency of nucleotide variants (NVs) in longitudinal samples from patient 1. Each longitudinal sample was sequenced once. Nucleotide variants (relative to reference sequence NC_045512.2) and potentially resulting amino acid mutations are indicated at the bottom or top of the map, respectively. Nucleotide variant C28628T only occurred in the background of C28629T, thus resulting in an A > F amino acid exchange in a fraction of C28629T mutations that alone led to an A > V exchange. The corresponding amino acid exchanges are marked with an asterisk. Variant frequency is indicated by heatmap colors, ranging from gray (reference) over yellow to dark blue, as indicated in the legend shown to the right. Ct values and treatment regimen are shown to the left. None of the investigated samples exhibited coverage levels below 10 (as labeled with a blue dot in Figures S2, S3, S4, and S5). Lower panel: Color code map indicating classification of nucleotide variants according to the observed coverage/frequency values, as further described in the STAR Methods section and illustrated in Figure S1. Classification calls corresponding to individual colors are shown in the legend to the right of the map (n.s.: not significant).